Positive Identification of Rhododendron through DNA Fingerprinting
A. Lane Rayburn, M.J. Iqbal and Donald W. Paden
University of Illinois at Urbana-Champaign
The capability of reliably identifying individuals, both humans and plants, by means of genetic fingerprinting is a recent breakthrough that has received considerable publicity in popular as well as scientific publications. In the plant world, techniques have now been developed that reveal genotypic distributions in blackberry and raspberry species and, in our own laboratory, in rye and wheat. Adapting these techniques to rhododendrons has the potential to resolve questions that frequently arise concerning whether specific plants are true representatives of a given species, whether a claimed cross has actually been achieved, or whether plants offered for sale are correctly identified.
A number of other important possible uses of DNA fingerprinting are obvious. A DNA fingerprint could be used when formally registering plants with the ARS and the Royal Horticultural Society. Tissue culture laboratories, after lengthy periods of propagating plants using the same culture, could check on whether genetic mutation had occurred. It also seems possible that the technique could be used in reducing the time and effort in finding partners for varieties that do not cross easily. For example, what is likely to accept pollen from the newest recognized member of the rhododendron family,
Rhododendron greenlandicum
(ledum) - or vice versa? The technique also might make it easier to estimate the relationships between cultivars so that breeders could introduce and maintain greater diversity while developing new varieties. The Rhododendron Species Foundation could perhaps maintain a file of "fingerprints" for important on-site plants. The list is almost endless.
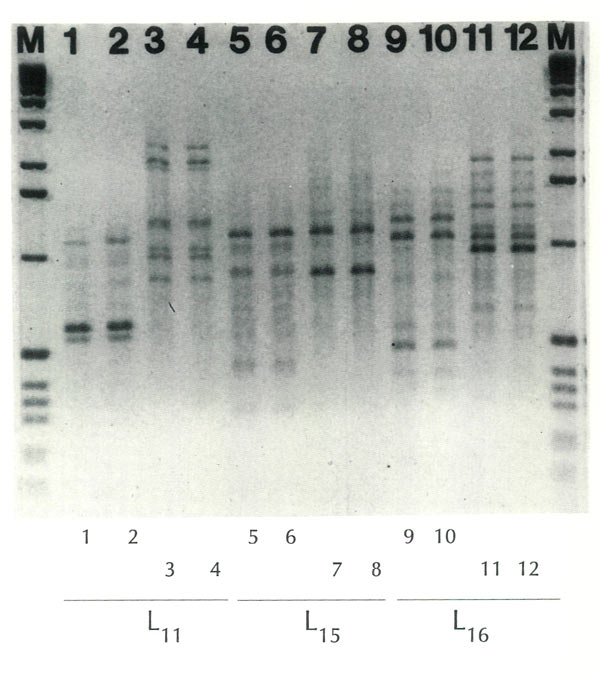
The nature of a fingerprint, or a profile, can be seen in Figure 1. Here profiles are displayed for two plants,
R. atlanticum
and
R. arborescens
.
1
At the bottom of this figure three "primers" are indicated, L
11
, L
15
, and L
16
. Each of these primers was used twice on each plant. It will be observed that the profiles for 1 and 2 are identical, as are 3 and 4, etc. The fact that each set of two is the same lends credibility to the procedure. The use of the same primer (e.g., L
11
) on material from
R. atlanticum
and
R. arborescens
results in obviously different prints (compare 1 and 2 with 3 and 4), thus providing a means of differentiating between the two species. Two additional primers were used (L
15
and L
16
), and each shows that the two plants are different.
|
|
Figure 1. Fingerprints of two rhododendron species with 3 primers (RAPDs).
M = Marker (1 Kb Ladder). Photographed in the cytogenetics laboratory at the University of Illinois, Urbana-Champaign; enlargement by the University Photographic Service. |
Arrays such as these are sufficiently complex to make possible identification of a very large number of different cultivars. It may also develop that such arrays offer the possibility of numerically describing plants so that they could be entered into a computer. Searches could then serve to identify identical or closely related plants in the same way that police departments search for a match with a human fingerprint obtained from a suspect.
2
The following detailed description of the technical procedures used to produce the above results is provided for readers who are interested in such information.
Procedures
After initial trials with several types of plant material, 11 plants were purchased from the Rhododendron Species Foundation in order to insure that the results of the experiment would represent true species. To isolate DNA from evergreen rhododendron species, different parts of the plant such as leaves (young and old), stem (including apical meristem), pollen, embryos, callus, and roots were tried.
3
After several unsuccessful attempts, DNA was isolated from very young leaves by cutting them from the plant and inserting them directly in liquid nitrogen. This probably inhibits the release of phenolic compounds which inhibit the isolation of good quality DNA that can be used for Polymerase Chain Reaction (PCR).
From deciduous rhododendron species, DNA isolation was much easier. Very young leaves were cut from the plant and dipped in liquid nitrogen, then ground into very fine powder. DNA was isolated by the method of Rogers and Bendich (1988) as follows. After grinding the tissue, equal volume of hot (65°C) 2X CTAB (2% cetyldimethyl-ethylammonium Bromide, 100 mM Tris [pH 8.0], 20 mM EDTA [pH 8.0], 1.4 M NaCl and 1% PVP [Polyvinyl-pyrolidone] Mr 4000) was added and kept at 65°C for 5 minutes. Then an equal volume of chloroform-isoamyl alcohol was added, mixed to form an emulsion and spun at 11000g for 30 seconds in a microfuge. The aqueous layer was removed and 1/10 volume of 10X CTAB (10% CTAB, 0.7 M NaCl) was added, mixed and then an equal volume of chloroform-isoamyl alcohol was added, mixed to form an emulsion and centrifuged as described above. The aqueous layer was removed and DNA was precipitated with a precipitation buffer: 1% CTAB, 50 mM Tris (pH 8.0), 10 mM EDTA (pH 8.0). After the solid DNA was centrifuged, it was re-suspended in high salt TE (Tris-EDTA) buffer and precipitated with 100% ethanol. After washing the pellet with 80% ethanol, the pellet was dried and re-suspended in .1X TE. Ribonuclease (RNAse) was added to the DNA solution to remove any ribonucleic acid (RNA) present.

|
DNA Fingerprinting
DNA fingerprinting was done by PCR using a Hybaid thermal reactor. DNA fingerprinting is based on the premise that plant species have different DNA composition. During the PCR reaction these small differences are amplified so that they are easily observed and compared. To DNA fingerprint a plant species, a small amount of DNA is added to a special fingerprinting mixture. In this mixture are enzymes and other chemical compounds which allow targeted DNA to be amplified. This amplification is based on sequence recognition. At 94°C, the two strands of the double stranded DNA molecule are separated. At 36°C, the primers (or target sequences) find their complementary sequences and then at 72°C, Taq DNA polymerase (DNA synthesizing enzyme) forms a new strand of DNA starting from the primer. In this way two molecules of a specific part of DNA are formed. These new molecules are then used to make additional molecules. The more cycles molecules undergo, the more DNA sequences are made. In this way, at the conclusion of the PCR process a large number of copies of a specific sequence are produced which can easily be detected by running the sample on an agarose gel and staining by Ethidium Bromide. Isolating the DNA and fingerprinting may take anywhere from two days to weeks.
The foregoing summarizes very briefly the work accomplished in this project. The goal was to develop an appropriate technique for securing profiles in the genus
Rhododendron
. This has been accomplished. Fingerprints were obtained from two evergreen rhododendron,
R. formosum
and 'Olga Mezitt', as well as from the two deciduous species reported above. These two profiles have not been included because it was felt that they would not contribute to the explanation just presented.
Future Research
In addition to what has been suggested above, profiles identifying other species and hybrids would be of interest to the profession and would demonstrate the usefulness of the technique. For example, fingerprints from two plants from different sources purporting to be the same species might be examined to ascertain the correctness of such a claim. Similarly, profiles of hybrids might be examined to study the reasonableness of their parentage. Profiles of parent plants might be compared with those of their progeny to determine whether there are differences in those derived from seeds, cuttings, or tissue culture.
Reference
Rogers, S.O., and Bendich, A.J., 1988. Extraction of DNA from plant tissue. In S.E. Gelvin and R.A. Schelperoort (Eds.),
Plant Molecular Biology Manual
, AG: 1-10, Kluwer Academic Publishers, Dordrecht, Netherlands.
1
Excellent photos of trusses of
R. atlanticum
and
R. arborescens
are featured in an article in the
ARS Journal
, Vol. 46, Summer, 1992, pp. 146-7. Both of these species are also shown and briefly described in
Horticulture
, Vol. 70, May, 1992, p. 54.
2
An interesting discussion of human DNA fingerprinting appears in the
National Geographic
, Vol. 181, May, 1992, pp. 102-123.
3
Pollen was furnished by James W. Gerdemann, tissue culture material was provided by Martin M. Meyer, Jr. and Don Paden, who also supplied several plants.
The research reported here was supported by a grant from the ARS Research Foundation.
A. Lane Rayburn is Assistant Professor and M.J. Iqbal is a Doctoral Candidate, Department of Agronomy, and Donald W. Paden is Professor Emeritus, Department of Economics, University of Illinois at Urbana-Champaign.