JARS 49n4 - Enzyme Fingerprinting of Rhododendron Cultivars
Enzyme Fingerprinting of Rhododendron Cultivars
Stephen L. Krebs
The David G. Leach Research Station of the Holden Arboretum
Abstract
Starch gel electrophoresis was used to identify rhododendron cultivars by distinguishing them on the basis of enzyme variability (isozymes) controlled by 10 genes. Of 61 Leach hybrids surveyed, 57 (93%) had unique isozyme fingerprints. Distantly related individuals showed the greatest differences, but the technique was also powerful enough to differentiate highly related plants such as cultivars from the same grex (full sibs). Among Dexter hybrids, three morphologically distinct variants of 'Scintillation' could not be distinguished by isozymes. However, two other potentially synonymous cultivars, 'Apritan'* and 'Dexter's Honeydew', proved to have different fingerprints. Isozymes were also used to compare the fingerprints of source cultivars with rooted cuttings taken from them. No variation was observed, indicating clonal fidelity during vegetative propagation. Similarly, micropropagated plants of 'Montego' and 'Lee's Dark Purple', including individuals with tissue-proliferation symptoms, were genetically identical to source plants for all isozymes examined. This suggests that rhododendrons propagated via tissue culture are also genetically stable.

|
Synopsis
Rhododendron enthusiasts are frequently vexed by problems of plant identification. Who has not speculated about the identity of an unlabeled rhododendron, wondered about the parentage of plants lacking ancestries, or been mystified by plants with the same clonal name that look slightly different?
Until recently, comparison of plant morphology and phenology has been the only method of determining like and unlike in the vast world of cultivated rhododendrons. This article describes an alternate approach to plant identification using the technique of isozyme analysis. Isozymes (iso-enzymes) are multiple forms of an enzyme which can be directly observed in the laboratory, and which vary among individuals and populations just as leaf or floral traits exhibit multiple forms (polymorphisms) in nature.
The research described here establishes that rhododendron cultivars from the Leach breeding program are readily identifiable by their unique enzyme "fingerprints." The fingerprinting approach was then extended to a pair of mini-studies, one examining the identities of some Dexter hybrids and the other looking into the question of clonal fidelity in rhododendrons propagated by cuttings and tissue culture.
Introduction
A nursery has several thousand cuttings of a rhododendron that appear to have been mislabeled. Can the identity of these plants be determined unambiguously without waiting for them to bloom? In cultivars such as 'Vulcan', 'English Roseum', and 'Scintillation', a group of variants with different morphology or phenology exist under the same name. Do these plants represent genetically different individuals, such as siblings or sports? A hybridizer notices that some seedlings of a cross have characteristics absent in either parent and suspects that the pollen source or seed lot may have been contaminated. Can the non-hybrid contaminants be positively identified and culled as seedlings? In another scenario, a patent is being developed for a Phytophthora resistant rhododendron that is morphologically similar to several other cultivars. Can some technique other than disease testing be used to distinguish the patented plant from look-alikes?
These situations raise questions of individual plant differences which can be resolved with genetic markers. Although most rhododendron cultivars can be readily identified at maturity based on their morphology and phenology, another set of distinguishing characters, such as biochemical or molecular markers, would be advantageous for several reasons. Whole-plant traits are not always sufficient to distinguish very similar cultivars, where subtle genetic differences may not be directly observable. Outward comparisons may also be confounded by the propensity of traits such as flower color, bloom date, leaf shape, and plant habit to vary in the same cultivar grown under different environmental conditions. In contrast, biochemical or DNA gene markers are less influenced by environmental variability and can be used to determine the genotype of a cultivar at any stage of development. This latter attribute is important because it can provide a timely answer to questions of genotypic differences. Current reliance on flower characters to differentiate rhododendrons can result in delays caused by seasonal changes or immature plants.
Although public awareness of genetic markers has been heightened most recently by the O.J. Simpson trial, these tools have been used extensively for the past two decades in plant breeding applications. Two types of markers, isozymes and DNA, are generally employed. Rayburn et al. (6) recently used DNA markers to distinguish two rhododendron accessions. The present study demonstrates the potential of isozyme markers for identifying and distinguishing among a large group of rhododendron cultivars. The technical aspects of visualizing and interpreting isozymes are briefly described first, followed by several research applications. The basic issues addressed in the research were: 1) how effective are isozymes in fingerprinting rhododendron cultivars? 2) can isozymes detect genetic differences among variants of a single cultivar or, conversely, confirm identity between two cultivars generally considered synonymous? and 3) do isozymes support the concept of genetic fidelity (cloning) in conventionally propagated and micropropagated plants?
Principles of Isozyme Analysis
The general principles and practice of isozyme analysis are presented below and in the sidebar on this page. Technical details about enzyme extraction procedures, gel and electrode buffers, and enzyme staining schedules used for rhododendrons are reported elsewhere (3).
Isozymes (iso-enzymes) are multiple molecular forms of an enzyme that catalyze the same substrate but differ in electrophoretic mobility (5). Through a technique called starch gel electrophoresis, enzymes are separated in a gelatinous starch matrix by running a strong electrical current through it. Because enzymes are proteins consisting of acidic, basic, and neutral amino acids, they migrate to different positions in the gel depending on their amino acid composition and net electrical charge. These differences in mobility (the distance traveled on the gel) allow us to visualize isozymes.
The basic experimental procedure is as follows. Total protein extracts from plant tissue (typically leaves) are inserted at the cathodal (-) end of the gel, electrodes are attached to it, and the power is turned on. Proteins are allowed to migrate towards the anode (+) for 4-5 hours. Out of the total array of proteins separated during this migration, a specific enzyme can be visualized by incubating the gel in a solution containing the substrate for that enzyme and color dyes which attach to the product formed by the enzymatic reaction. Isozymes then can be seen as colored bands with different mobilities on the gels, some migrating further towards the anode than others ("fast" vs. "slow" isozymes).
Enzymes are usually the products of single genes. Formal genetic studies of the genes controlling enzymes are greatly aided by some of their user-friendly features: Mendelian inheritance, codominant expression, and much less environmental influence than for morphological traits (7). A recent study of hybrid elepidote rhododendrons demonstrated that variability observed in nine enzyme systems is controlled by 15 different genes, and that isozyme variants (alleles) produced by these genes are inherited in a Mendelian fashion (3). As a result of this research, the number and relative mobilities of isozyme bands produced by different plants can be interpreted as genetic fingerprints.
| Table 1. Fingerprints of 61 Leach hybrids (54 elepidotes and 7 lepidotes) at 10 genes controlling enzymes† | ||||||||||
| CULTIVAR†† | ENZYME | |||||||||
| Aco -1 | Aco -3 | Idh -1 | Mdh -1 | Mdh -2 | Mdh -3 | Pgd -1 | Pgd -2 | Pgi -2 | Pgm -2 | |
| 'Last Hurrah' | aa | aa | cc | dd | bb | ab | cc | bb | ce | bd |
| 'Ballad' | ab | aa | cc | cd | bb | bb | cc | bb | cc | bb |
| 'Spellbinder' | ab | aa | ce | cd | bb | bb | cc | bb | ce | bb |
| 'Summer Snow' | ab | ae | cc | cd | bb | ab | cc | bb | ce | bb |
| 'Edmond Amateis' | ad | aa | ce | cd | bb | bb | cc | bb | cc | bb |
| 'Ceylon' | ad | ac | cc | cd | bb | ab | cc | bb | cc | bb |
| 'Nepal' | ad | ac | cc | cd | bb | bb | cc | bb | ac | bb |
| 'Bravo!' | ad | ac | ee | cd | bc | ab | bc | bb | cc | bb |
| 'Vee Vee' | bb | ce | dd | bb | bb | bc | bb | ce | bb | |
| 'Trinidad' | bb | aa | cc | cd | bb | bb | cc | bb | cc | bd |
| 'Shenandoah' | bb | aa | cc | cd | bc | bb | cc | bb | cc | bb |
| 'Boule de Rose' | bb | aa | cc | dd | bb | bb | cc | bb | cc | bb |
| 'Sumatra' | bb | aa | cc | dd | bc | ab | cc | bb | cc | bb |
| 'Small Wonder'(3) | bb | aa | ce | cd | bc | bb | cc | bb | cc | bb |
| 'Crete' | bb | ac | ac | cc | be | bb | cd | ab | ce | bb |
| 'Tow Head' | bb | bd | aa | bc | ab | bb | ae | bd | ce | dd |
| 'Malta' | bb | bd | dg | ab | bd | bb | ac | bb | ee | de |
| 'Ivory Coast' | bb | be | df | bb | bb | bb | ac | bb | ef | ce |
| 'Hudson Bay' (9) | bb | dd | af | bb | ab | bb | cc | be | cf | de |
| 'Siam' | bd | aa | ac | cd | be | bb | ac | ab | cc | bb |
| 'Samoa' | bd | aa | ac | dd | bb | bb | bc | bb | cc | bb |
| 'Burma' | bd | aa | cc | bc | bb | cc | bb | cc | bb | |
| 'Nile' | bd | aa | cc | cd | bb | bb | cc | bb | cc | bb |
| 'Rangoon' (3) | bd | aa | cc | dd | bb | bb | cc | bb | cc | bb |
| 'Singapore' (3) | bd | aa | ce | cd | bb | bb | cc | bb | cc | bb |
| 'Canary Islands' | bd | aa | ce | cd | bb | bb | cc | bb | cc | bb |
| 'Tennessee' | bd | aa | cf | cd | bb | bb | cc | bb | cc | bb |
| 'Spring Frolic' (5) | bd | ac | ac | cd | be | bb | cc | ab | cc | bb |
| 'Lodestar' (8) | bd | ac | cc | dd | bb | ab | cc | bb | ce | bb |
| 'Luxor' | bd | ac | cc | dd | bb | bb | cc | bb | cc | bb |
| 'Bangkok' (4) | bd | ac | ce | cd | bb | bb | cc | bb | ce | bb |
| 'Hindustan' | bd | ac | ce | dd | bb | bb | bc | bb | cc | bb |
| 'Bali' (7) | bd | ac | ee | cd | bb | bb | cc | bb | cd | bb |
| 'Monaco' (4) | bd | ac | ee | cd | bb | bb | cc | bb | ce | bb |
| 'Cyprus' (1) | bd | ac | ee | dd | bb | bb | cc | bb | ce | bb |
| 'Red River' | bd | ae | cc | cd | bb | ab | cc | bb | ce | bb |
| 'Summer Glow' | bd | ae | cc | dd | bb | bb | cc | bb | cc | bd |
| 'Senegal' | bd | bd | bd | bb | ab | bb | ac | be | cc | bd |
| 'Jericho' | bd | be | ad | bb | ab | bb | ac | bc | ce | cc |
| 'Nuance' (7) | bd | cc | de | cd | bb | bb | cc | bb | ce | bb |
| 'Duet' (4) | bd | cc | ee | cd | bb | bb | cc | bb | cd | bb |
| 'Yukon' (9) | bd | dd | bf | bb | ab | bb | cc | bd | bf | de |
| 'Summer Solace' | bd | ee | cc | bb | bb | cc | bb | ce | bb | |
| 'Casanova' | cd | aa | cc | dd | bc | bb | cc | bb | cc | bb |
| 'Normandy' (6) | cd | aa | cc | dd | bc | bb | cc | bb | cc | bb |
| 'Montego' | cd | aa | dd | dd | bc | ab | cc | bb | cc | bb |
| 'Swansdown' (8) | cd | ac | cc | dd | bb | ab | cc | bb | cc | bb |
| 'Persia' (1) | cd | ac | cc | dd | bb | bb | cc | bb | cc | bb |
| 'Party Pink' (1) | cd | ac | ce | dd | bb | bb | cc | bb | cc | bb |
| 'Ravenna' | dd | aa | ac | cd | bc | bb | cc | bb | cc | bd |
| 'Hong Kong' | dd | aa | cc | cd | bc | ab | cc | bb | cc | bb |
| 'Borneo' | dd | aa | ce | bb | ab | cc | bb | ac | bb | |
| 'Rio' (6) | dd | aa | ce | cd | bb | bb | cc | bb | cc | bb |
| 'Anna H. Hall' (5) | dd | ac | aa | cd | bc | bb | cc | ab | cc | bb |
| 'Great Lakes' (5) | dd | ac | ac | cd | bc | bb | ac | ab | cc | bb |
| 'Ivory Tower' | dd | ac | cc | cd | bb | bb | cc | bb | ac | bb |
| 'Golden Gala' | dd | ac | cc | dd | bc | bb | cc | bb | cc | bb |
| 'Applause' (2) | de | aa | dd | cd | bb | bb | cc | bb | ce | bb |
| 'Finlandia' (2) | de | aa | dd | dd | bb | bb | cc | bb | cc | bb |
| 'Robin Leach' (2) | de | aa | dd | dd | bb | bb | cc | bb | ce | bb |
| 'Flair'(2) | de | ac | dd | dd | bb | bb | cc | bb | ce | bb |
|
† Conventional isozyme notation is used to designate enzymes, genes, and variants (alleles) produced by those genes. The 6 enzymes used for this study were aconitase (ACO), isocitrate dehydrogenase (IDH), malate dehydrogenase (MDH), 6-phosphogluconate dehydrogenase (PGD), phosphoglucoisomerase (PGI), and phosphoglucomutase (PGM). The enzyme-controlling genes are designated in italics, followed by a number (-1, -2, or -3); the lowest number indicates the fastest-migrating isozyme (mostanodal) and higher numbers refer to products of slower-migrating genes if more than one gene is present. As indicated above, ACO, MDH, and PGD are encoded by multiple genes in rhododendrons, a frequent occurrence in many other plant species (7). Enzyme variants at each gene are shown as letters indicating relative mobilities; a is the fastest-migrating variant, b the next fastest, and so forth. Pairs of letters refer to the diploid (sporophytic) genotype. Homozygotes have the same letter pair and heterozygotes have different letters. †† Cultivars followed by the same number in parentheses represent full-sibs (grexes). Highlighted rows represent pairs of cultivars that could not be separated on the basis of their fingerprints. Sorting of cultivars was done in alphabetically ascending order on the basis of isozyme variant designations, starting with Aco -1 and sorting at subsequent genes in the order given above. |
||||||||||
Results and Discussion
Issue 1: Can isozymes be used to establish unique genetic fingerprints for cultivars?
The population chosen for this study included 54 elepidote and seven lepidote cultivars produced by David G. Leach (Table 1). Many of these cultivars are inter-related to some degree because of pedigree selection and the repeated use of certain cold-tolerant clones (such as 'Catalgla') as parents. These 61 plants included nine grexes (groups of full-sib cultivars) and several half-sib cultivars. Thus the power of isozyme analysis to discriminate among individuals varying widely in relatedness could be tested.
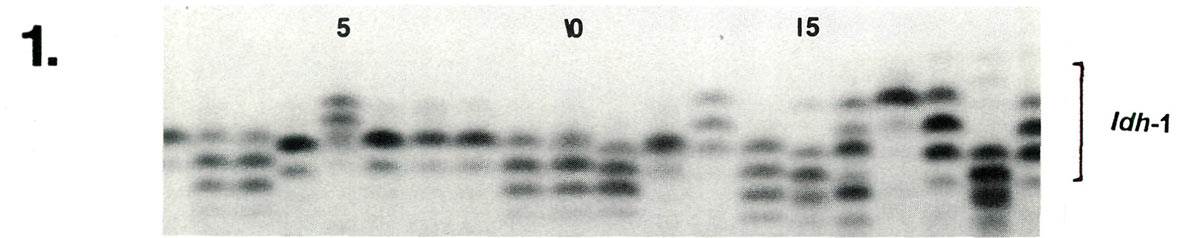
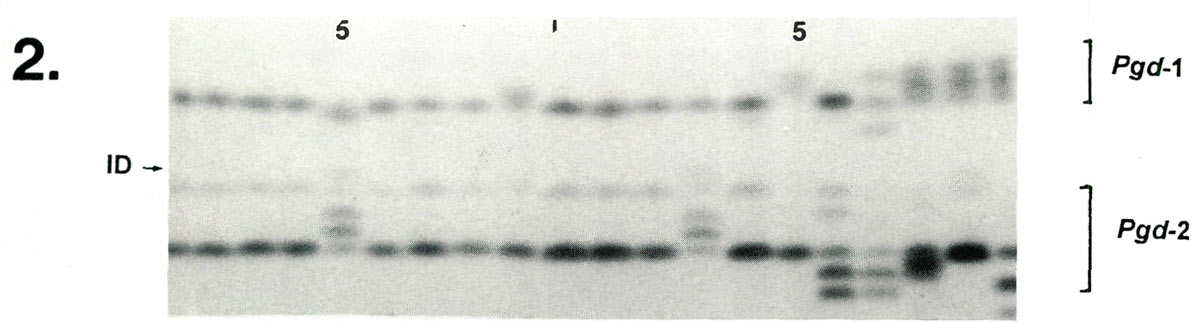
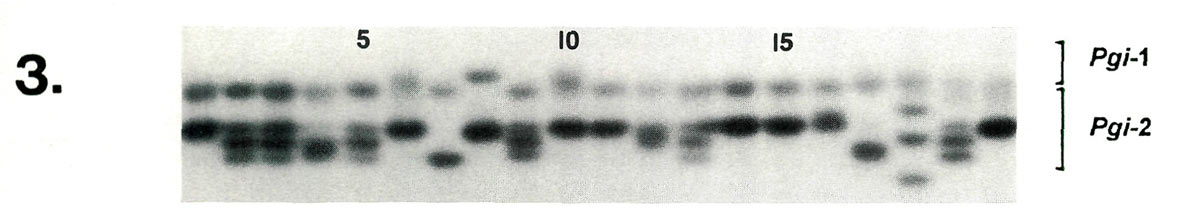
Examples of enzyme variability among cultivars are given in Figures 1-3 along with genetic interpretations of the gels. The results of the completed cultivar screen are given in Table 1, which lists the variants alleles) present in the 61 cultivars at 10 enzyme-controlling genes. Starting with the first gene ( Aco -1), the cultivars are sorted by alleles (labeled a, b , c , ....) in alphabetically ascending order. Two alleles are assigned to each gene in accordance with the probable diploid chromosome of these cultivars.
Fifty-seven out of 61 (93%) cultivars were shown to have unique fingerprints by this method. All members of grexes (full-sibs) were clearly separated on the basis of isozyme differences, although they often differed by only one variant at a single gene, such as the 'Applause', 'Findlandia', 'Robin Leach', and 'Flair' grex. This result is consistent with their high degree of relatedness. Limitations of the fingerprinting technique were observed in two cases where pairs of cultivars could not be distinguished (highlighted in Table 1). The fact that 'Casanova' and 'Normandy' are half-sibs may explain why they were identical at all 10 sampled genes. More inexplicable is the isozyme similarity of 'Singapore' and 'Canary Islands', which are morphologically distinct and not closely related.
| Table 2. Enzyme variability among 54 elepidote and 7 lepidote Leach hybrids 1 | |||
| Gene | Variants shared by elepidotes and lepidotes | Variants unique to elepidotes | Variants unique to lepidotes |
| Aco -1 | b, d | a, c, e | -- |
| Aco -3 | e | a, c | b, d |
| Idh -1 | a, d, f | c, e | b, g |
| Mdh -1 | c | d | a, b |
| Mdh -2 | b | c, e | a, d |
| Mdh -3 | a, b | -- | -- |
| Pgd -1 | a, c | b, d | e |
| Pgd -2 | b | a | c, d, e |
| Pgi -2 | c, e | a, d | b, f |
| Pgm -2 | d | a, b | c, e |
| 1 Summarized from Table 1. | |||
It should be noted that although both lepidote and elepidote cultivars shared isozymes in common (Table 2), each subgeneric group contained unique enzyme variants not observed in the other. This is most likely a reflection of the evolutionary past of the constituent species. At some point the lepidote and elepidote species shared a common ancestor, but once they diverged and became reproductively isolated any new isozymes resulting from mutations became unique to that group. In oversimplified terms, this demonstrates how isozymes can be used for taxonomic and phylogenetic purposes, applications that have been carried out for many other plant genera (2).
|
| Lane | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| Idh -1 | cc | ce | ce | dd | ac | cc | cc | cc | ce | ce | de | cc | ac | cf | dg | bf | aa | ad | df | bd |
|
| Lane | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| Pgd -1 | cc | cc | cc | cc | cd | cc | cc | cc | bc | cc | cc | cc | cc | cc | ac | cc | ae | ac | ac | ac |
| Pgd -2 | bb | bb | bb | bb | ab | bb | bb | bb | bb | bb | bb | bb | ab | b | bb | bd | bd | be | bb | be |
|
| Lane | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| Pgi -2 | cc | ce | ce | de | ce | cc | ee | cc | ce | cc | cc | cd | ce | cc | cc | cc | ee | bf | ce | cc |
| Key to Figures 1-3: Listed below are the cultivars and accessions corresponding to lanes 1 - 20 in the gel photographs. | ||||||||
| Lane | Figures 1 & 2 | Figure 3 | Lane | Figures 1 & 2 | Figure 3 | Lane | Figures 1 & 2 | Figure 3 |
| 1 | 'Rangoon' | 'Burma' | 9 | 'Vee Vee' | 'Monaco' | 17 | Tow Head' | 'Malta' |
| 2 | 'Small Wonder' | 'Red River' | 10 | 'Canary Islands' | 'Siam' | 18 | 'Jericho' | 'Yukon' |
| 3 | 'Singapore' | 'Summer Summit' | 11 | 'Nuance' | 'Bikini Island' | 19 | 'Ivory Coast' | 'Tow Head |
| 4 | 'Finlandia' | 'Blazen Sun' | 12 | 'Swansdown' | 'Peach Parfait' | 20 | 'Senegal' | 'Senegal' |
| 5 | 'Crete' | 'Red Sea' | 13 | 'Spring Frolic' | 'Spellbinder' | |||
| 6 | 'Summer Snow' | 'Spring Frolic' | 14 | 'Tennessee' | 'East Grove Yellow'* | |||
| 7 | 'Scarlet Blast' | 'Mount Mitchell' | 15 | 'Malta' | R. oreodoxa var. fargesii | |||
| 8 | 'Summer Glow' | R. degronianum ssp. heptamerum | 16 | 'Yukon' | 'Vulcan' | |||
Issue 2: Is the current recognition of cultivar variants and synonymous cultivars supported by isozyme analysis?
Synonymous cultivars are those with different names that appear to be identical in the garden. Cultivar variants share a common name, but look slightly different in morphology and/or phenology. Both occur among rhododendron hybrids. Isozyme analysis was applied to some of these cultivars to determine whether current groupings and separations based on whole-plant characteristics could be substantiated.
Dexter hybrids were used for this study. Three variants of 'Scintillation' were supplied by Briggs Nursery, the so-called "Mehlquist", "eastern," and "western" types, which differ slightly in bloom date and leaf morphology according to their descriptions in the 1994 Briggs catalog. These were compared with three older plants of 'Scintillation' from the East Coast, including one from the Heritage Plantation, supplied by J. Leonard. Also included was a pair of Dexter hybrids, 'Apritan'* and 'Dexter's Honeydew', which are considered to be synonymous (J. Leonard, personal communication).
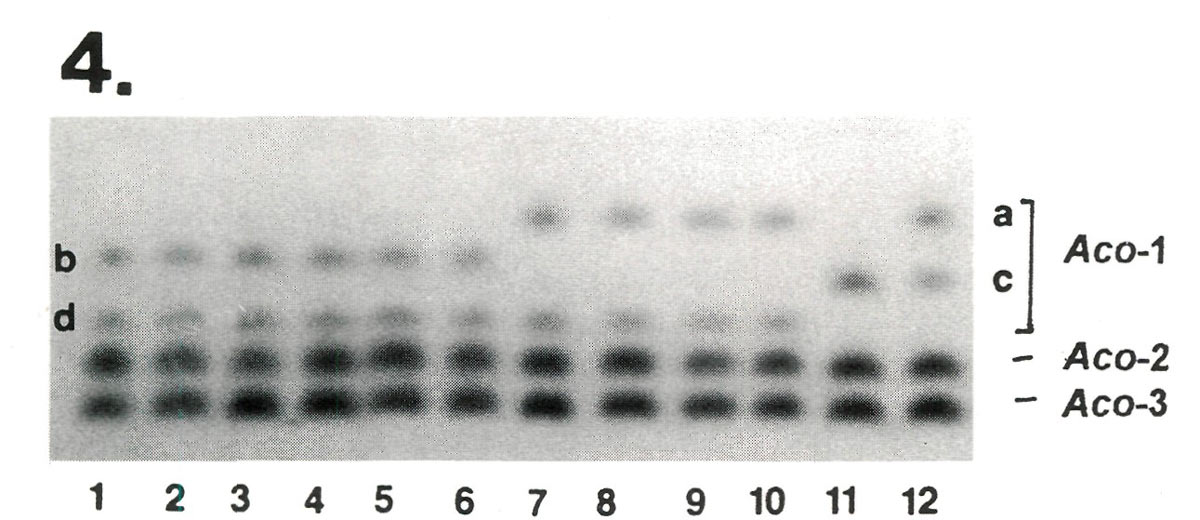
After electrophoresis, protein extracts from these plants were stained for seven enzymes comprising 13 genes. The results from one of the enzymes systems are shown in Figure 4. No fingerprint differences were observed among the variants of 'Scintillation' in any of the enzymes sampled. However, 'Apritan'* and 'Dexter's Honeydew' exhibited different isozymes at the Aco -1 gene (Fig. 4), indicating that they are not identical.
|
|
Figure 4. Comparison of Dexter hybrids at 3 genes controlling variability in aconitase (ACO), a monomeric enzyme.
Aco -2 and Aco -3 are invariant, while Aco -1 varies among cultivars. Lanes 1-6 represent the 3 variants of 'Scintillation' from Briggs Nursery plus 3 older plants of 'Scintillation' from the East Coast. All plants were identical bd heterozygotes at Aco -1. The 'Scintillation' samples proved to be indistinguishable for all other enzymes tested as well. In contrast, 'Apritan'* (lane 11) and 'Dexter's Honeydew'(lane 12) exhibited different genotypes at Aco -1 ( cc vs ac ), indicating that they are not synomous. [Note: The Dexter material was not compared on the same gel with Leach hybrids. Therefore, these designations a, b, c, and d are made without reference to the isozyme variants described in Table 2] |
These results demonstrate some of the powers and limitations of genetic markers. Based on a single difference, markers can unambiguously demonstrate that two individuals are different (e.g., 'Apritan'* and 'Dexter's Honeydew'). However, when markers do not differ among plants, it can only be said that it is likely but not certain that they are genetically identical (e.g., the 'Scintillation' variants). This is because the 13 isozyme markers sampled represent only a small portion of the total genome, and individuals may differ at other unsampled genes. The fingerprints of 'Canary Islands' and 'Singapore' described in the previous section exemplify the caution that should be exercised when drawing conclusions from these data. Based on isozyme data alone, one would be led to the erroneous conclusion that the two cultivars are identical.
At present, the current recognition of three 'Scintillation' variants is not supported by the isozyme data. Screens of the Dexter material with additional enzyme or DNA-based markers might reveal differences not detected here, but there is no guarantee that this will occur. The subtle variations observed in leaf morphology or bloom date among 'Scintillation' plants may reflect single gene differences derived from somatic mutation (sports) or open-pollination of the source cultivar. Enzyme or DNA markers linked to such differences might be found, but locating them would require much time and expense, as well as a bit of luck. To test whether the 'Scintillation' variants are genetically different, it might be simpler to make crosses between them and observe whether leaf morphology, bloom date, or other traits segregate as discrete characters among the progeny. Self-pollinated progeny from each form could be included in this experiment as check populations.
Issue 3: Do isozymes confirm that propagation is a clonal process?
Successful vegetative propagation requires that the product be genetically identical to the source. This cloning process is achieved either by rooting cuttings or by promoting shoot proliferation from meristems in tissue culture. In rare instances, mutations occur in somatic tissue which can give rise to cuttings that are genetically different from the original material. An opportunity to check for clonal fidelity in rhododendron propagation exists at the David G. Leach Research Station, where the original clone of each cultivar is maintained along with multiple copies grown from rooted cuttings. At least five different plants of 'Cyprus', 'Edmond Amateis', 'Lodestar', 'Nepal', 'Party Pink', 'Senegal', 'Spring Frolic', and 'Vernus' were compared for their enzyme banding patterns at 10 genes. Within each cultivar, the original and propagated plants exhibited identical fingerprints, evidence that vegetative propagation in rhododendrons is clonal.
The possibility of genetic changes during in vitro propagation has been raised in connection with the phenomenon of tissue proliferation (TP) observed in some rhododendron cultivars (1, 4). Do the gall-like symptoms appearing on the crowns of certain micropropagated plants derive from somatic mutations which have occurred during tissue culture? Isozymes were analyzed to test for this possibility. Two cultivars prone to TP were used in the experiment - the original clone of 'Montego' and a 15-year-old vegetatively propagated clone of 'Lee's Dark Purple'. Each was compared to 30 micropropagated plants of which approximately half were exhibiting TP symptoms (galls and/or adventitious shoots at crowns). After staining for eight enzymes (14 genes), intra-cultivar comparisons revealed no isozyme differences between the source clone and the micropropagated plants.
Within the limits of current isozyme technology for rhododendrons, these results suggest that tissue culture is a clonal process even where TP is occurring. TP symptoms may therefore result from epigenetic factors (transient gene expression) rather than from stable genetic changes induced by tissue culture conditions (1). If this is the case, changes in culturing protocols might result in a reduction of TP. At the same time, it would be wise to compare micropropagated populations using additional genetic markers to provide a more robust test of clonal stability. Research is currently in progress in other laboratories to help resolve these issues.
Isozyme Bands
The number of isozyme bands which appear on a gel depends on several factors: 1) the number of genes present 2) genotypic condition (heterozygous or homozygous) and 3) the number of polypeptide subunits which unite to form a functional enzyme. Information about the number of genes and enzyme variants (alleles) is obtained by progeny testing. Because isozymes are codominantly expressed, parents exhibiting different bands and carrying different alleles will produce F 1 hybrid progeny that show both parental bands (see below). The simplest case is for monomeric enzymes, where the product of each allele (a polypeptide subunit) represents a functional enzyme, and heterozygotes appear as double bands. For dimeric and tetrameric enzymes, heterozygotes exhibit additional bands which do not appear in either parent. These intermediate bands represent hybrid forms of the enzymes (heteromers) created when subunits encoded by different parental alleles are assembled into functional enzymes. In the dimeric system shown below, the F 1 heterozygote (ab) displays three bands, two homodimers formed from subunits produced by a single allele (aa or bb) and one heterodimer (ab) of intermediate mobility produced by each of the two parental alleles. For tetrameric enzymes, five bands appear in heterozygotes; two homotetramers (aaaa, bbbb) and three heterotetramers (aaab, aabb, abbb). The enzymes used in this study were either momomers or dimers.
A schematic illustration of idealized relationships between banding patterns, enzyme subunit number, and the genotypic condition of the enzyme-controlling gene. The crosses depicted involve parents homozygous for different isozymes, a heterozygous F 1 , and a segregating F 2 .
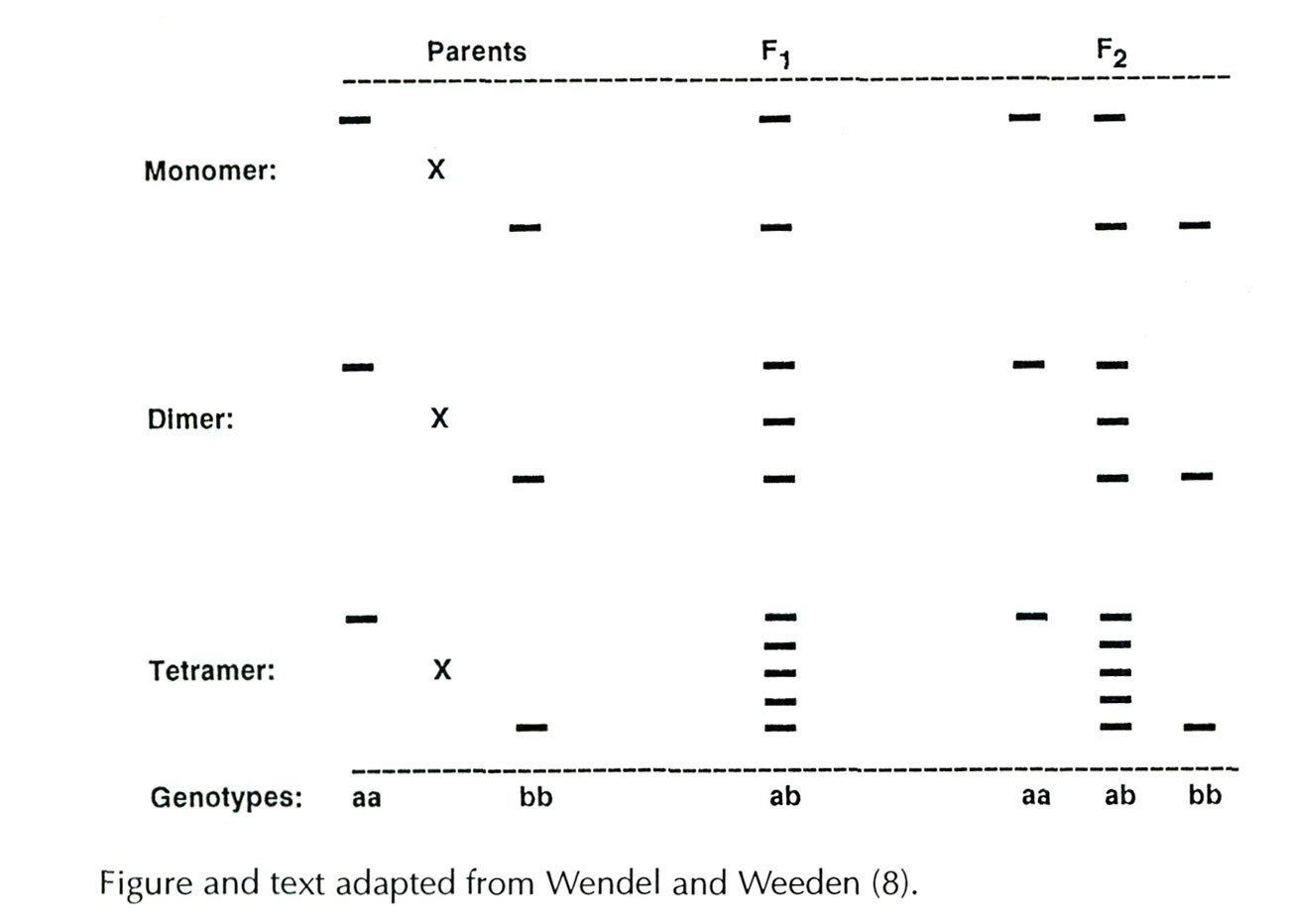
|
Acknowledgments This research was supported by a grant from the Research Foundation of the American Rhododendron Society.
References
1. Brand, M.H. 1992. Tissue culture variations: problems and solutions. Amer. Nurseryman 174(5): 60-71.
2. Crawford, D.J. 1989. Enzyme electrophoresis and plant systematics. In D.E. Soltis and P.S. Soltis (eds), Isozymes in plant biology, 146-164. Dioscorides Press, Portland, Oregon.
3. Krebs, S.L. Normal segregation of allozyme markers in complex rhododendron hybrids. J. Heredity (in press).
4. Linderman, R.G. 1993. Tissue proliferation. Amer. Nurseryman 178(5): 56-67.
5. Markert, C.L. and F. Moller. 1959. Multiple forms of enzymes: tissue, ontogenetic and species specific patterns. Proc. Natl. Acad. Sci. USA 45: 753-763.
6. Rayburn, A.L. and D. Paden. 1993. Positive identification of rhododendron through DNA fingerprinting. J. Amer. Rhod. Soc. 47: 137-138.
7. Weeden, N.F. and J. F. Wendel. 1989. Genetics of plant isozymes. In D.E. Soltis and P.S. Soltis (eds), Isozymes in plant biology, 46-72. Dioscorides Press, Portland, Oregon.
8. Wendel, J.F. and N.F. Weeden. 1989. Visualization and interpretation of isozymes. In D.E. Soltis and P.S. Soltis (eds), Isozymes in plant biology, 5-45. Dioscorides Press, Portland, Oregon.
* Name not registered.
