JARS v56n4 - Mapping Cold Hardiness Genes in Rhododendrons: An Assessment of Strategy
Mapping Cold Hardiness Genes in
Rhododendrons: An Assessment of Strategy
Rajeev Arora
Department of Horticulture
Iowa State University, Ames, Iowa
Stephen L. Krebs
The David G. Leach Research Station of the Holden Arboretum
Madison, Ohio
Chon C. Lim
Soft Landscaping
Jaya, Malaysia
Synopsis
Many molecular techniques and genetic strategies are available to hunt for genes controlling plant
traits. This paper describes our experiences with a gene-mapping approach called "bulked segregant
analysis." This strategy can provide a highly efficient means of detecting associations (linkage)
between hundreds or thousands of random DNA markers and the trait of interest. In our study, no
DNA markers with potential linkage to cold hardiness were discovered. A single marker with possible
linkage to the freeze sensitive phenoytpe was detected.
Introduction
Several years ago, we described a genetic study of cold hardiness in rhododendrons
(1)
that was
funded in part by the ARS Research Foundation. This study involved populations derived from a
Rhododendron catawbiense
x
R. fortunei
cross, in which progeny segregated for cold
hardiness, measured as leaf-freezing tolerance. Our genetic analysis of F2 and backcross seedlings
from this cross led to several conclusions: gene action involved in cold hardiness is additive
(little or no dominance involved), the cold hardiness trait in rhododendrons is under multigenic
control, and as few as three "key" genes could account for most of the phenotypic variation among
hardiness genes. Here we report our results from the preliminary stages of bulked segregant analysis,
and give an assessment of usefulness of this approach in our work. In addition, alternative
strategies for identifying cold hardiness genes are discussed.

|
Materials and Methods
Most of the technical background concerning DNA extraction and generation of randomly amplified
polymorphic DNA (RAPDs) has been omitted, but is available from the authors by request. The primers
used for generating RAPD markers via polymerase chain reaction were obtained from a commercially
available kit of 200 high GC content 10-mer oligonucleotides (UBC, Canada). DNA isolation and
polymerase chain reaction conditions followed the protocols of Marquard et al.
(3)
.
The conceptual aspects of combining RAPD marker generation with bulked segregant analysis are interesting and worth reviewing in more detail. In today's molecular world, mapping is often accomplished by finding random DNA sequences that are physically adjacent (linked) to the gene(s) of interest and therefore co-segregate with them during meiosis, in contrast to the independent segregation of unlinked genes characterized first by Gregor Mendel. With the advent of the polymerase chain reaction, the production of multitudes (thousands) of DNA markers (RAPDs) randomly distributed throughout plant genomes has become commonplace. Mapping with this multitude of random DNA segments is called a "shotgun" approach, since it is analogous to covering a broad area (the total plant genome) with a spray of pellets (markers) in the chance that one or more might hit the target (a specific gene). The most problematic and time-consuming aspect of mapping this way arises in trying to determine which, if any, of the many RAPDs have hit the mark.
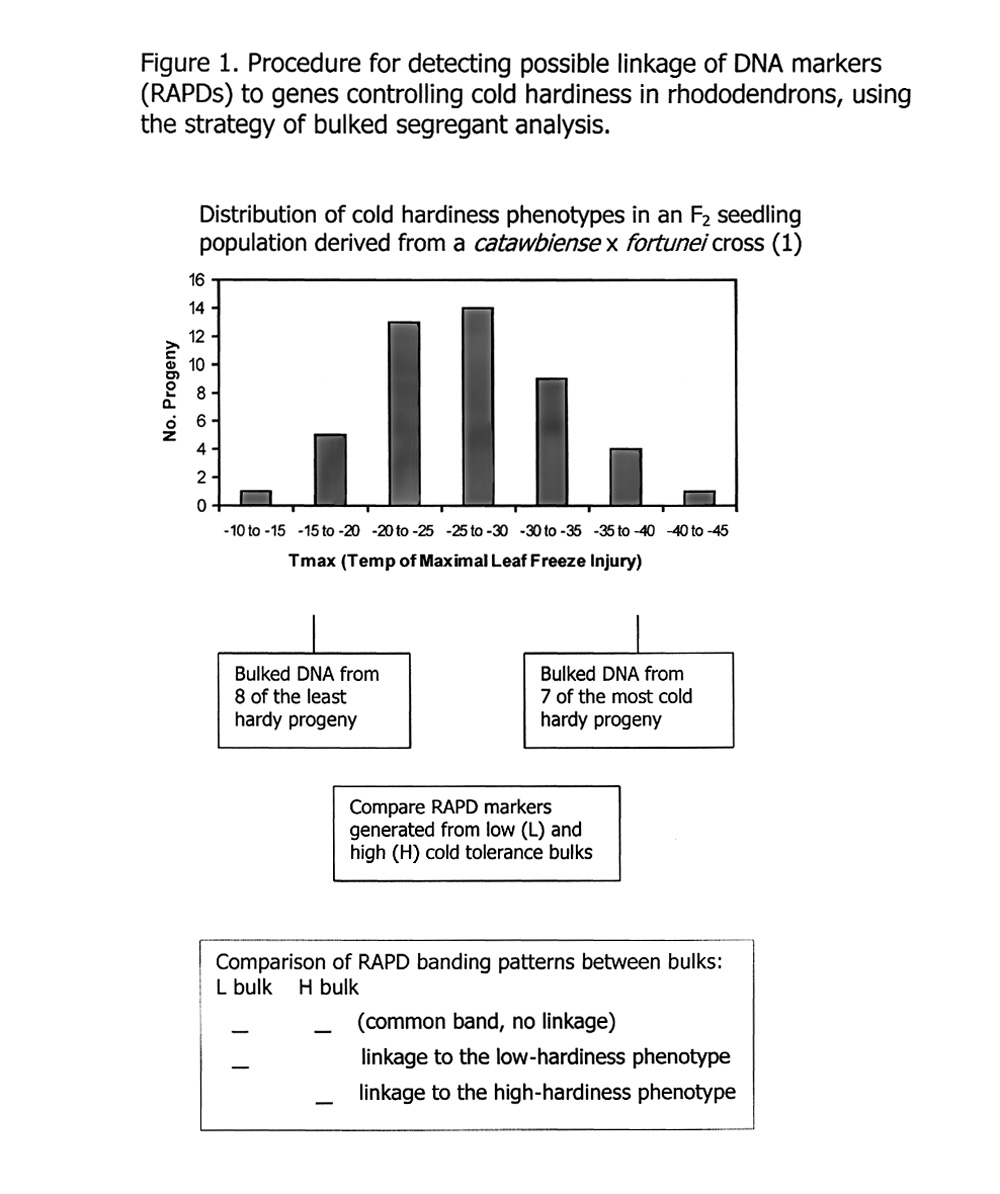
Bulked segregant analysis (Figure 1) offers a very efficient method for screening these markers and singling out those with potential linkage to a genetic trait. The method operates by identifying phenotypic "extremes" in populations (e.g., male vs. female, susceptible vs. resistant, most cold hardy vs. least cold hardy), collecting a few progeny within each extreme class, physically pooling (bulking) DNA from progeny in each group, and then comparing numerous DNA banding patterns in each bulk. One looks for DNA bands that appear in one bulk but are absent in the opposing phenotype. These bands are then identified as putative markers that may be physically linked on the chromosome to a gene controlling the trait of interest. The efficiency gained by using bulked segregant analysis is due to the fact that all markers are screened initially against a pair of bulked DNA samples rather than against many separate DNA samples from each individual in the population - a huge savings in time and expense.
For our study, we used progeny from an F2 population segregating for cold hardiness (1) , and selected eight progeny from the low-freezing-tolerance "tail" of the F2 distribution, and seven progeny from the high-freezing-tolerance tail of the distribution (Figure1). This resulted in two bulks in which the progeny were separated by an average 14.7°C in cold hardiness phenotype. Pooled DNA from each bulk was used as a template to generate RAPDs from the 200 primers. The random DNA fragments generated by polymerase chain reaction were separated and visualized on agarose gels, and the DNA banding patterns were interpreted as described above and in Figure 1 for possible linkage to cold hardiness.
|
Results
After the first screen, most primers showed no DNA banding pattern difference between the bulks. A total
of eighteen primers yielded bands displaying a presence versus absence difference between "low" and
"high" cold-hardy (CH) bulks, but when these were screened a second time using varying concentrations
of DNA, only four of the eighteen primers looked promising. After screening these four primers against
the DNA from individual progenies comprising the bulks (non bulked DNA), no patterns were observed in
which a RAPD band occurred exclusively in one set of progeny and not the other. The most consistent
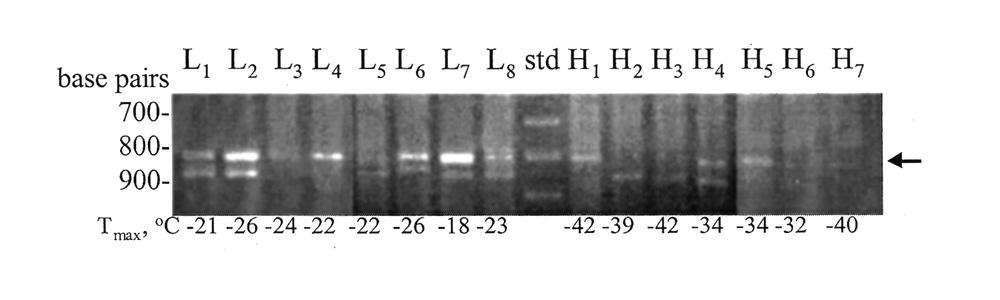
marker pattern derived from UBC primer #29, which yielded a 800-bp band in all of the eight "low-CH"
progenies, while three of the seven of the "high-CH" progenies had this band (Figure 2).
|
|
Figure 2. DNA profiles of individual F
2
plants after PCR amplification with primer
No. 29.
Lane 9 was loaded with 100 base pair (bp) ladder. L and H correspond to the "low" and "high" freeze-tolerant plants. T max = quantitative measure of leaf freezing-tolerance. |
Discussion
The screening process described above represents only the first stage in mapping via bulked segregant
analysis. Once putative markers are identified, they must then be screened using the full array of
segregants, not just the "extreme" phenotypes. This allows a quantitative estimate of the linkage to
be made, which describes the proximity of the DNA marker to the gene of interest as "strong" (close)
or "weak" (relatively far) linkage. Strong linkage of a marker is always desirable, since it means
that it usually segregates with the trait. The marker can then be used to track or predict the trait
in genetic or breeding studies, much as inherited diseases are traced in humans through genetic
testing.
The results of initial screenings indicate that further mapping with our set of markers is not warranted. At best, the 800-bp marker from primer UBC #29 is weakly linked to a gene conferring low cold hardiness. Although it is found in all the "low-CH" progeny, its presence in three of seven "high-CH" progeny suggests that recombination (crossovers) with a gene conferring high cold hardiness is occurring fairly frequently. High recombination rates indicate weak linkage; at a 50% rate the traits are unlinked.
Although we had anticipated finding markers linked to genes from the super hardy R. catawbiense parent, a converse result - linkage to relatively cold sensitive genes from R. fortunei - is not unexpected. From a practical standpoint, such as using markers to predict cold hardiness and assist in making selections in a breeding program, either outcome can be useful. In our study, an individual carrying the 800-bp marker is about two times more likely to be freezing-sensitive than freezing-tolerant.
Bulked segregant analysis has been successful in mapping many plant genes, particularly in cases where the trait being studied is monogenic (e.g., male vs. female, yellow vs. white flowered). The strategy can also work for mapping multiple gene traits, such as some forms of disease resistance in plants (5, 6). In our research, additional sets of primers (hundreds more are available) might yield useful RAPD markers, and use of other types of DNA markers could prove fruitful in mapping cold hardiness. Our results might also improve by continued testing of our F2 progeny to ensure that we have correctly classified leaf freezing phenotypes. Our estimates of cold hardiness were made on juvenile seedlings (~3 years old), and cold hardiness may change (increase) with age (2) . Studies of the F2 population at physiological maturity might change cold hardiness rankings and the composition of the bulks, leading to different results from marker analysis.
The RAPD mapping method is essentially a "shotgun" approach to understanding the genetics of cold hardiness. Many random DNA loci are generated with some likelihood that a few will be linked to genes of interest. Other methods, such as the "candidate gene" approach, may prove more fruitful. Over the past decade, researchers have identified numerous plant proteins that accumulate during cold acclimation and may play a role in stabilizing cell functions during freezing events. These proteins, often called cor (cold responsive) or dehydrins (induced by dehydration stresses such as freezing), have very similar properties and structures regardless of plant source. Recently, we have characterized differences in dehydrins among our F2 segregants, and have found that both genotypic- and age dependent differences in cold hardiness are associated with varying levels of a 25 kDa dehydrin from R. catawbiense (2) . Because this seems to be a more fruitful avenue for research at present, we have put the mapping project on hold and are investigating the role of rhododendron dehydrins in cold hardiness.
Acknowledgements
Journal paper No. 19694 of the Iowa Agriculture and Home Economics Experiment Station, Ames, Project No.
3601, and supported by Hatch Act and State of Iowa funds. This research was supported, in part, by a grant
from the Research Foundation of the American Rhododendron Society, West Virginia University Agricultural
and Forestry Experiment Station, and "in-house" funds from the Holden Arboretum.
Literature Cited
- Lim, C.C.; Krebs, S.L.; Arora, R. Genetic study of freezing tolerance in Rhododendron populations: implications for cold hardiness breeding. J. Am. Rhod. Soc. 52: 143-148; 1998.
- Lim, C.C.; Krebs, S.L.; Arora, R. A 25 kD dehydrin associated with genotype- and age-dependent leaf freezing tolerance in Rhododendron : a genetic marker for cold hardiness? Theor. Appl. Genet. 99: 912-920; 1999.
- Marquard, R.D.; Davis, E.P.; Stowe, E.L. Genetic diversity among witchhazel cultivars based on randomly amplified polymorphic DNA markers. J. Amer. Soc. Hort. Sci. 122: 529–535; 1997.
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. 88: 9828-9832; 1991.
- Molnar, S.J.; James, L.E., Kasha, K.J. Inheritance and RAPD tagging of multiple genes for resistance to net blotch in barley. Genome 43: 224-231, 2000.
- Xu, M.L.; Melchinger, A.E.; Xia, X.C.; Lubberstedt, T. High-resolution mapping of loci conferring resistance to sugarcane mosaic virus in maize using RFLP, SSR, and AFLP markers. Mol. Gen. Genet. 261: 574-581, 1999.
Dr. Rajeev Arora (corresponding author; rarora@iastate.edu ) is an Associate Professor of Horticulture in the Department of Horticulture at Iowa State University, Ames, IA. Dr. Stephen Krebs is a plant breeder and Chong Lim, a former doctoral student in Dr. Arora's research program, is a Project Manager at Soft Landscaping, a subsidiary of CyPark Sdn Bhd, in Petaling Jaya, Malaysia.
