JARS v60n1 - Regional DNA Variation within Rhododendron macrophyllum
Regional DNA Variation within
Rhododendron macrophyllum
Zhenxiang Xi, Amy Denton, Shermi Liang, Laura Sugden, Matt Hodson,
and Benjamin Hall
University of Washington
Seattle, WA
Rhododendron macrophyllum , the Pacific coast rhododendron, is considered to be a single species. It has a range extending approximately from San Francisco to Victoria and from the Pacific Ocean inland to the Cascade crest (Clark, 1999). Throughout this region, its distribution is very patchy, with continuous large populations only in the rain shadow of the Olympic Mountain range and in several parts of Oregon (WNARSP, 2005). Previous studies of variation within R. macrophyllum (Boge, 1986, 1988) have mainly dealt with flower color, which can vary from dark pink to white. To test for more subtle differences among plants from different populations, we have been analyzing the DNA of R. macrophyllum leaves and flower buds over the last 10 years. Researchers on this project included, at various times, high school sophomores, college juniors and an advanced graduate student. The results lead to the conclusion that Pacific coast rhododendron populations contain several genetically distinct types. As botanists, we are eager to determine how these differences arose. For horticulturists, it would be useful to know how these types differ from one another in important traits, such as resistance to summer drought, and to be able to identify each of them for hybridization purposes.

|
|
The photograph of
Rhododendron macrophyllum
is by Dennis Hendrickson, who
found the plant on the Tahuya-Blacksmith Road approximately 6 miles west of Bremerton, WA, in Kitsap County. The plant was growing in a logged area that had been replanted. Approximately 90 percent of the samples in this area have clade 1 genes. |
In 1995 when Amy Denton, a botany graduate student at the University of Washington, began her doctoral research in molecular systematics she chose to focus on Rhododendron . Her general plan was to isolate a segment of DNA from the same genomic location either in many individual plants of the same Rhododendron species or in many different species and then to determine for each the sequence of A's, G's. T's and C's in the segment. Although living collections of many species were available at the Rhododendron Species Foundation garden in Federal Way, WA., near Seattle, the results of her initial experiments led her to concentrate on a single species, R. macrophyllum . Surprisingly, for the DNA region Amy studied, there was more variation from plant to plant in the R. macrophyllum population than there was between species in other Hymenanthes taxa (Denton, 1997).
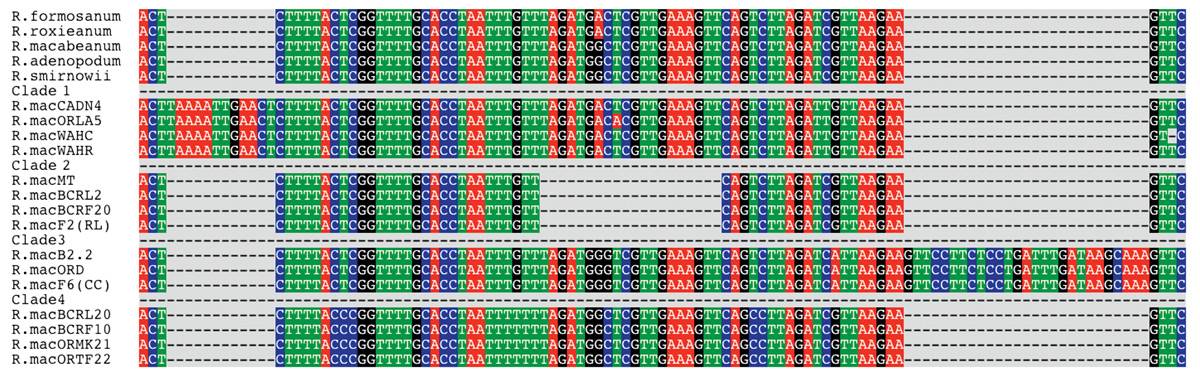
Viewing directly the DNA region that Amy sequenced (Figure 1), two features leap to the eye: sequences for the five species at the top, even though they represent different subsections within subgenus Hymenanthes , are extremely similar to one another. For Asian Hymenanthes species, differences were only seen in the case of R. degronianum and R. makinoi , which share a common insertion within this region.
On the other hand, sequences of the R. macrophyllum gene, from samples representing twenty plants taken at various locations, can be separated into four distinct categories. We refer to these categories as clades. A clade is a group of organisms that are thought to share a common ancestor not shared with other clades. When Amy completed her thesis research, the data suggested that certain clades were geographically localized (Denton, 1997). For example clade 1, found in California, Oregon and Washington, generally was found near sea level, while clades 3 and 4 tended to be present at higher elevation sites in the interior. This correlation was not perfect and the total number of samples was only 30, yet there was a definite trend, inviting further study. That did not occur until several years later.
In 2003, a teacher-mentor relationship was established between Laura Sugden, science teacher at West Seattle High School (WSHS), and Ben Hall, Professor at the University of Washington. Rhododendron DNA experiments were carried out by the students in their classroom, with supplies and equipment provided by the Science Education Partnership at the Fred Hutchinson Cancer Center in Seattle. Flower-bud samples for DNA analysis were collected on student trips to R. macrophyllum populations not sampled in Amy Denton's research. In 2003-2004, the class collected specimens at three locations: the Mountaineers Theater near Bremerton, WA, Mount Mitchell (South of Mt. St. Helens) and Cypress Island, near Anacortes, WA. The last two of these collection trips, particularly the Cypress Island one, gave the students a taste of the adventure that naturalists and plant collectors relish.
In the words of Laura Sugden: "West Seattle High School is a large inner city school where there are a high number of students who have not had many outdoor experiences. In fact for some of them, these collection trips were some of their first experiences with the wilderness. Any project in school with hands-on experience increases the level of excitement and enthusiasm. These trips and subsequent processing of the plant extract converted many of my students to amateur botanists."
Hien Lu, a student who went on all of the trips, writes, "The experience I got by going on these outings with Dr. Hall has been great! When I went to Cypress Island, 'boy', that was a walk. It was raining cats and dogs, and the hike was straight up. I felt like I was in the shower! I remember we finally found the rhododendrons and cheered. We had to climb down the steep side of the hill to get to them. They were all growing on a slanted hill. At one point of the walk I wanted to give up, but after we found the plants it was worth it! After we finished the walk we collapsed down on the beach while waiting for our private boat to get us. I loved the boat ride. I saw eagles for the first time that day. I loved it, and will never forget it."
Again, quoting Laura Sugden: "My students and I have been honored to take part in real-life research. In my opinion, it is these types of experiences that are most successful at promoting a career in the sciences to our young people."
DNA sequences of the WSHS collected samples from Mount Mitchell and Cypress Island, and of samples collected at Shawnigan Lake, near Victoria B. C., by Don Whittle fell nicely into place with Amy Denton's data. Cypress Island and Shawnigan Lake samples were in clade 1 (maritime), and the Mount Mitchell sample was in clade 2, which seems to occur throughout the region. These results, together with Amy's, left little doubt that, across the range of R. macrophyllum in the Northwest, there were regional genetic differences, suggesting the need for an in-depth study. Collecting trips and DNA analysis by Laura's class continued into the next year. In order to increase the pace of research, additional researchers were recruited.
The University of Washington and Sichuan University in Chengdu, China, have a junior-year student exchange program involving 25 to 30 students per year. Research on the natural environment and on human impact are the general themes of the program, and one specific area is the study of biodiversity. The biodiversity students coming from Sichuan have a strong interest in learning how to use genetics and DNA analysis to measure plant diversity. When they are told that the State of Washington has only five Rhododendron species, including several Ledum and Menziesia species, they find this very surprising. A comparable area of Sichuan would have more than 50 Rhododendron species (Chamberlain et al . 1996). What Washington lacks in species diversity is, however, partially made up by the DNA diversity of R. macrophyllum (Figure 1).
|
|
Figure 1. Aligned DNA sequences of Asian Rhododendron species and individual
R. macrophyllum
plants. The DNA sequence is that of intron four of the
RPB2 gene.
The dashed line areas represent sequence gaps (absence of DNA) in the positions shown. |
For the R. macrophyllum DNA project, a decision was made to collect and sequence samples from many more individual plants and to do this at widely separated locations. This was accomplished by several collecting trips with students in the UW-Sichuan program, supplemented by samples collected in various parts of Oregon and northern California by Wilber Bluhm, Jason Ashford, Mike Oliver and Dick Cavender and then mailed to the University of Washington. The first large-scale collection in 2004/5 was made along the Canadian arm of the Skagit River, between Ross Lake and Rhododendron Flat on Highway 3. For this, Professor Gerry Allen of the University of Victoria guided a group that included Dr. Hall, Shermi Liang, a UW biology graduate, and five recently arrived students from Chengdu. The ride was long and bumpy and the border formalities daunting, but the Skagit rhodies were spectacularly healthy plants, full of flower buds which were harvested and preserved in silica gel for DNA extraction. Shermi and Zhenxiang then became the backbone of the R. macrophyllum project for the next nine months. Over the winter, intensive specimen collecting was done on the Kitsap Peninsula, Olympic Peninsula, and Whidbey Island, and a very interesting specimen was obtained from Detroit in the central Oregon Cascades.
When the sequencing of these DNA samples was completed, the sequences aligned and interpreted, Amy Denton's inferences about the R. macrophyllum population structure were resoundingly confirmed. All copies of the gene sequenced (which we call intron 4) belonged to one of the four clades. All of the Puget low-land sites contained primarily clade 1, with a sprinkling of 2, and the upper Skagit rhodies belonged primarily to clades 2 and 4, with some clade 1. We decided that, before completing the project, we should consult the R. macrophyllum experts, members of the Western North America Rhododendron Species Project (WNASRP). This group has constructed and published maps of where R. macrophyllum occurs (Clark, 1999, 2002).
In February, 2005, Xi, Liang, Hall, Sugden and high school students Hien Lu and Olivia Sengsi made a presentation of their results to a group consisting of WNARSP members Helm, Clark, Dunning, O'Neill and Peterson, as well as Sonja Nelson, Estella Leopold, Arthur Kruckeberg, Toby Bradshaw and Andrew Eckert. There was a high level of enthusiasm to continue and complete the project, and specific advice was given to sample the large population of R. macrophyllum along the central Oregon coast. These would be expected to have a high content of clade 1 genes, if the hypothesis of an ancestral maritime distribution were correct. Also, plans were made for Laura Sugden's 2004-2005 science class to accompany Hank Helm on an expedition to Wind River, in the Columbia Gorge, to collect R. macrophyllum specimens there for DNA analyses in the classroom and at the University of Washington.
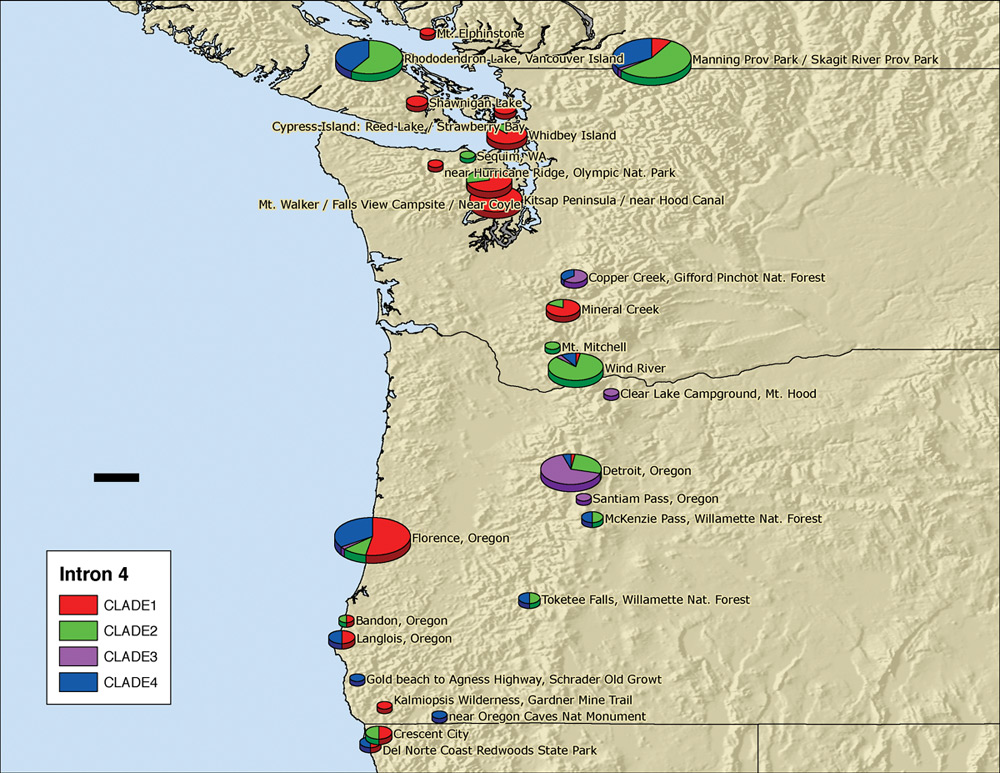
As the 2004-2005 school year drew to a close, major expeditions to collect R. macrophyllum were made to Rhododendron Lake on Vancouver Island, to Wind River and, finally, to Florence, Oregon. Zhenxiang, Shermi, Ben, Clarice Clark and Jerry Broadus made the first expedition immediately after the ARS Convention in Victoria. The samples obtained from Rhododendron Lake and Wind River added strength and substance to the database, while specimens collected in and near Florence filled a major gap in the geographic coverage. Additional foliage samples were sent to us from Oregon by Bluhm and Ashford and from California by Cavender and Oliver. During June and early July, exceptionally productive DNA analysis by Zhenxiang determined the DNA clade for each of these new samples. At that time, Shermi's job of placing DNA sequencing information on the map (Figure 2) was taken over and completed by Yunmi Kim, a graduating UW senior majoring in biology.
|
|
Figure 2. Genotypic composition of
R. macrophyllum
populations in the
Pacific Northwest. At locations where more than one
sequence type was present, some diploid plants were heterozygous. Therefore, the scoring of clades 1 through 4 on which this figure is based, counts homologous chromosomes of each type, not plants of each type. |
All together, over the period from 1995 to 2005, DNA samples from 250 R. macrophyllum plants and from 40 plants of related species have been analyzed for their intron 4 DNA sequence. As each specimen was collected, the location of the plant was determined by a GPS (global positioning system) reading and entered in the field notebook. All samples were taken in silica gel to the University of Washington, where their DNA was extracted and their intron 4 genotype was determined by DNA sequencing. Knowing the latitude and longitude of each sample site from GPS and the genotype of the plant (clade 1, 2, 3 or 4), we entered the data points on the map (Figure 2). The relation of colors to the various clades is shown on the left side of the map.
Because quantization is essential, multiple samples were taken and analyzed for each of the populations recently studied. On the map (Figure 2), the size of each pie is proportional to the number of samples from that local region. For example, the data at the Skagit River site represent samples from 35 plants. The relative widths of different pie slices show the proportion of each clade at that site. The results in Figure 2 show that, genetically, R. macrophyllum is markedly non-uniform across its native range. The concentration of red at coastal sites indicates that clade 1 is generally found near salt water; these locations would tend to have the least extreme summer and winter temperatures. Many, but not all of these, are in low-precipitation (rain-shadow) areas. Clade 3, indicated by purple, is the main type found in the central Oregon Cascades. Copper Creek in Washington is the only other site at which it was found to be abundant. Clades 2 and 4 are more widespread and are the principal forms at mountainous sites in British Columbia and southern Oregon.
Developing a hypothesis to explain our DNA data (Figure 2) has required an interweaving of findings from paleo-botany, geology, atmospheric science, molecular evolution and plant systematics. First we inferred, from the differences in DNA composition at different sites, that long ago the ancestors of these populations separated and followed different migration paths. After they became reproductively separated, their DNAs diverged. How long ago might that separation have occurred? Fossilized rhododendron leaf impressions from Republic, Washington, date to the mid-Eocene (50 million years ago [mya]) (Wehr and Hopkins 1994). The overall mixture of fossil flora there, at Princeton, B.C., and at an Idaho site clearly indicate the presence of an upland forest, with moderate moisture levels in the interior Northwest during and after the Eocene period. Fossil leaf and pollen specimens suggest that, throughout the Miocene (24 to 5 mya), rhododendrons were present in eastern Washington and other parts of the Northwest. Contrast this to the modern distribution of R. macrophyllum (Clark, 1999). None of it is present on the western slopes of the Olympic Mountains or the Washington Cascades north of Mt. Rainier. No R. macrophyllum is found east of the Cascades or in the Canadian Okanagan. In Southern Oregon, the coastal and Cascade Range populations are continuous with one another but further north, they are separated by the Willamette Valley, which has no native R. macrophyllum .
Major geological changes explain most of the differences in the occurrence of Rhododendron between the present and the Miocene. The formation of the Cascades 5 mya and lava flows in the Columbia Basin rendered much of Washington state unsuitable habitat for Rhododendron survival, since the western slopes became too wet and eastern Washington subsequently experienced very dry summers (Leopold and Denton, 1987). As these changes occurred, range reduction just east of the rising Cascades may have happened gradually, with the eastern populations managing to persist at a few semi-moist sites near the Cascade crest and possibly mixing to some extent with R. macrophyllum from the west side. This may account for the prevalence of clade 4 genes on the upper Skagit River in British in the southern Oregon mountains, and in the Columbia Gorge. Clade 3 may also have come from east of the Cascades for, with the exception of Copper Creek, it is now only found in abundance near the Cascade crest.
During the Pleistocene, glaciers must have subjected the surviving rhododendrons of the Pacific Northwest to even greater hardships. Most of the rain shadow areas of the western half of the Puget basin were covered by an ice sheet many hundreds to thousands of feet thick as recently as 16,000 years ago during the Fraser glaciation (Porter and Swanson, 1998). This created the rocky glacial deposits of the southern Kitsap Peninsula, on which R. macrophyllum now thrives. The indications are that the ancestors of these rhododendrons persisted through the glacial maximum in a small refugium on the Olympic Peninsula and then repopulated the Olympic and Kitsap Peninsulas (Buckingham et al., 1995). The small size of the surviving founder population would explain the DNA uniformity of present-day R. macrophyllum on the Olympic and Kitsap Peninsulas, Whidbey and Cypress Islands and lower Vancouver Island. Virtually every plant sampled in this large area belongs to clade 1. Pleistocene glaciers very likely affected R. macrophyllum in parts of Oregon, as well. The great Lake Missoula floods that resulted from the breakup of successive ice dams that backed up the Clark Fork River were responsible for flooding and silting the Willamette Valley. Unlike the hilly terrain and well-drained soils west of Puget Sound, the fine-grain sediments deposited in the Willamette Valley led to a low-relief landscape ideal for growing grass, but not R. macrophyllum .
The ecological and climatic history of the Northwest may indeed provide an explanation for the present distribution of clades 1, 2, 3 and 4 throughout the region. When Rhododendron populations suffered range reduction over the last several million years, genes of other (some of them now extinct) species may have been incorporated into R. macrophyllum . A specific case in point concerns the intron 4 sequence of clade 4. An alignment and sequence comparison was made between R. macrophyllum clade 4 sequences and those of other rhododendron species either closely related to R. macrophyllum ( R. aureum , R. catawbiense , and R. brachycarpum ) or overlapping in range with R. macrophyllum ( R. occidentale ). Out of these and other intron 4 sequences in our database, the ones most resembling R. macrophyllum clade 4 are found in R. occidentale . This raises the possibility of clade 4 having entered the R. macrophyllum genome by introgression; that is, as the result of a natural hybridization event between ancestors of R. occidentale and ancestors of R. macrophyllum with subsequent back-crossing to other R. macrophyllum plants. This would be analogous, but not identical, to the cross that led to 'Oregon Queen', a sterile artificial hybrid.
We are often asked: "does this type of research have any practical consequences?" In fact, it may have. Our interpretation of the colored ovals in Figure 2 is that R. macrophyllum populations around the Hood Canal and nearby islands and in the central Oregon Cascades are relatively homogeneous, genetically. On the other hand, those around Florence, Oregon, are genetically very mixed, including various hybrids between three different ancestral types (clades 1, 2 and 4). Because there are genetic differences between populations from different locations and because many of the populations have withstood selection under various adverse conditions, it follows that highly favorable combinations of genes may be present at certain locations, genes that confer resistance to various types of environmental stress. These might include the ability to withstand low winter temperatures (upper Skagit and central Oregon Cascade populations), to grow in very wet conditions (Rhododendron Lake, Wind River), or to survive summer drought (Olympic Peninsula, Hood Canal), to name just a few. Together with WNARSP members, we are now considering the possibility of planting R. macrophyllum test gardens at several locations, choosing plants from various sites to test on the basis of their genetic makeup. Accompanying this would be intensive study of other regions of the R. macrophyllum genome, on which studies are being initiated (Goetsch et al., 2005).
Regardless of whether we can achieve these and other future goals toward understanding R. macrophyllum , our project has been well worth the effort. Sixty high school students and a half dozen college juniors have had experience with DNA in the lab and with rhododendrons in the wild. We hope that for a number of them the experience has influenced their outlook and their prospects.
Acknowledgement
We thank all of those mentioned in this article, as well as Keith White and Gene Cockeran, for their assistance. Without the network of very helpful people who make up the American Rhododendron Society, this research could not have been carried out.
References
Boge, D. 1986. The Ross-Boge
Rhododendron macrophyllum
expedition.
J. Amer. Rhod. Soc.
40(2) 62-65.
----------1988. The Ross-Boge
Rhododendron macrophyllum
expedition - 1987.
J. Amer. Rhod. Soc.
42(3) 162-167.
Buckingham, N. M., E. G. Schreiner, T. N. Kaye, E. Burger, and
E. L. Tisch. 1995. Flora of the Olympic Peninsula. Northwest
Interpretive Association, Seattle, WA. Pp 20-38.
Chamberlain, D. E, R. Hyam, Argent, G. Fairweather, and K. S.
Walter. 1996.
The genus Rhododendron its classification
and synonymy
. Edinburgh: Royal Botanic Garden.
Clark, C. 1999. Mapping
Rhododendron macrophyllum
in
the Wind River area of Washington state.
J. Amer. Rhod.
Soc.
53(2): 99-103.
---------- 2002. Mapping
Rhododendron macrophyllum
on Whidbey and Cypress Islands, Washington state.
J.
Amer. Rhod. Soc.
56(1): 2-6.
Denton, A. 1997. The Evolution of RNA Polymerase II
Introns: Ancient Polymorphism and Paraphyly in the
Genus
Rhododendron
(Ericaceae). Doctoral
Dissertation, University of Washington.
Goetsch, L., A. J. Eckert, and B. D. Hall. 2005.
The molecular systematics of
Rhododendron
(Ericaceae): a phylogeny based upon RPB2 gene
sequences.
Syst. Bot.
30(3): 616-626.
Leopold, E. B. and M. F. Denton. . 1987. Comparative age
of grassland and steppe east and west of the
northern Rocky Mountains.
Ann. Miss. Bot. Gard.
74: 8941-867.
Porter, S. C. and T. W Swanson. 1998. Radiocarbon age
constraints on rates of advance and retreat of the
Puget lobe of the Cordilleran ice sheet during the last
glaciation.
Quaternary Research
50: 205-213.
Wehr, W. C. and D. Hopkins. 1994. The Eocene orchards
and gardens of Republic, Washington.
Washington Geology
22(3): 27-54.
Western North American Rhododendron Species Project. 2005.
