JARS v50n2 - DNA Fingerprinting of Rhododendrons: Phase II
DNA Fingerprinting of Rhododendrons: Phase II
Donald W. Paden, M. Javed Iqbal, A. Lane Rayburn
University of Illinois at Urbana-Champaign
Synopsis
This is the second of two articles on the use of DNA fingerprinting to identify rhododendrons and to quantify their differences. The first article (Rayburn, Iqbal & Paden, 1993) was focused on the process of obtaining fingerprints. In this one, DNA fingerprints are presented for a number of different well-known rhododendrons, for several named hybrids, and for seedlings from a single plant. The possible usefulness of this technique is discussed.
The process of obtaining DNA fingerprints of rhododendrons was explained in an earlier Journal article (Rayburn, Iqbal, & Paden, 1993), where results were illustrated with fingerprints of two azaleas, Rhododendron atlanticum and R. arborescens . Fingerprints for two small-leafed (lepidote) rhododendrons were also obtained and mentioned in the article, but were not illustrated. Additional research and the extension of this procedure to other rhododendrons now make possible a better understanding of its importance.
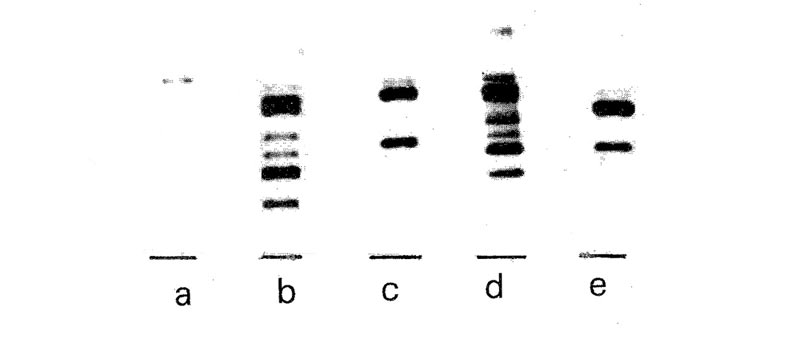
The five fingerprints shown in Figure 1 represent several major categories of rhododendrons along with Ledum groenlandicum , referred to in this article as R. groenlandicum . Ledum groenlandicum is included as a possible member of the genus Rhododendron (see Kron & Judd, 1990; also Anonymous, 1991). All of these profiles were obtained from fresh leaves, with the exception of 'Olga Mezitt', where tissue-cultured shoots were used. The DNA fingerprints of these plants differ widely from each other.
|
|
Figure 1. DNA fingerprints of five rhododendrons, (a). Large leaf: 'Roseum Elegans',
(b). Small leaf: 'Olga Mezitt', (c). Azalea: R. arborescens , (d). Vireya: ( R. lochiae x R. pseudonitens ) x R. commonae , (e). R. groenlandicum . |
The next set of prints (shown in Fig. 2) is of four additional plants. It is important to note that here two prints are shown for each plant. (This duplication is ordinarily done in the laboratory to show that if the procedure is duplicated the prints will be the same.) 'Mist Maiden' (labeled c) and 'Ken Janeck' (labeled d) are two named clones of R. yakushimanum and are almost identical, as would be expected. The first set of prints, R. smirnowii x R. yakushimanum , are of a hybrid and would not be expected to be the same as the prints for the two clones just described. On the other hand, 'Pink Parasol' is registered as a named clone of R. yakushimanum (see Salley & Greer, 1992). The prints for our plant, purchased with the 'Pink Parasol' label, are noticeably different from 'Mist Maiden' and 'Ken Janeck'. All of this leads to the observation that a possible application of the fingerprinting procedure would be in settling disputes as to the correct identification of plants for sale or in the classification of plants for other purposes. Flowers of 'Pink Parasol' and R. smirnowii x R. yakushimanum are shown in the accompanying photographs.

|
|
Figure 2. DNA fingerprints of four plants involving
R. yakushimanum
. (a).
R. smirnowii
x
R. yakushimanum,
(b). 'Pink Parasol',
(c). 'Mist Maiden', (d). 'Ken Janeck'. Photographs compare the first two plants. |
The third set of prints, shown in Figure 3, are from six seedlings of 'Adele's Yellow' ( R. maximum x R. wardii ), selfed, grown from the same parent plant. The fingerprints are very different. If these profiles had come from plants propagated from the mother plant asexually, they would have been the same, with only relatively minor variations (see Rouse et al., 1993). The photograph shows the six plants used to obtain fingerprints from the seedlings of 'Adele's Yellow'. The plants differ somewhat from each other in appearance: two have noticeably red stems and the others differ slightly in their growth habit. A photograph of the flower of the mother plant is also included.

|
| Figure 3. Fingerprints of six seedlings of 'Adele's Yellow', selfed. |
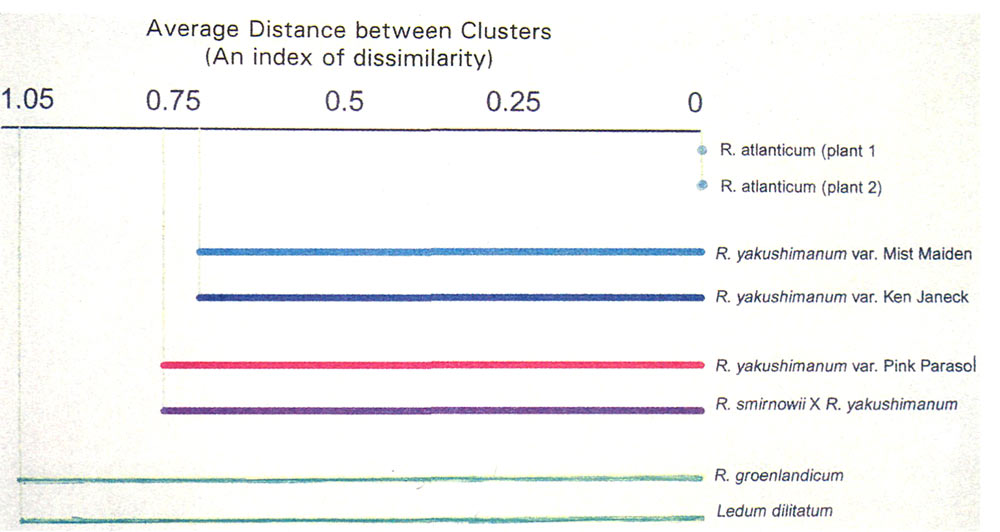
The results of another analysis of the data are presented in Figure 4 (p. 98), with the plants arranged in pairs solely for purposes of discussion. An index was computed showing the dissimilarity of the eight plants in question compared with a group of 13 rhododendrons which were used overall for comparison. These included one azalea, six elepidotes, three lepidotes, one vireya, and two ledum. (See last paragraph of the Technical Addendum, following; also Rayburn et al., 1993. Not all of the plants in this database are mentioned specifically in this article.)
The computer was asked how dissimilar the two forms of ledum are to the plants in the database. (Note: our Ledum dilatatum was purchased with this name, although it may not be correct.) The indexes for these two plants, for all practical purposes, are the same. Their indexes differed very much, however, from all of the rhododendron plants in the database. The difference in their genetic composition is undoubtedly related to the difficulty of crossbreeding R. groenlandicum with other rhododendrons.
At the other extreme, the dissimilarity index for the two plants of R. atlanticum (propagated asexually from the same plant) shows no difference. The indexes comparing these plants with others in the database is very low (i.e., they have much in common with the other plants).
|
|
Figure 4. Cluster analysis of RAPD (Randomly Amplified Polymorphic DNA)
amplification profiles produced from various rhododendrons. |
Perhaps of greater interest is the comparison of the indexes for the middle four plants in Figure 4. The index for the two named clones of R. yakushimanum , 'Mist Maiden' and 'Ken Janeck', are very similar to each other (but differ considerably from all others in the database). On the other hand, the dissimilarity index for our 'Pink Parasol' differs from the pair just mentioned and is closer to the index of the hybrid R. smirnowii x R. yakushimanum . This supports our previous question of whether our 'Pink Parasol' plant may be a hybrid.
A file of fingerprints of the plants at the Rhododendron Species Foundation would provide a unique service to those interested in rhododendrons. A generous benefactor willing to finance the startup costs of such an undertaking might make the project possible. Also required would be the continued cooperation of a laboratory, probably at a teaching institution, with suitable equipment and a willingness to provide fingerprints (appropriately priced) for members of the American Rhododendron Society, the Rhododendron Species Foundation, and others. It would be helpful, of course, if DNA fingerprints of rhododendrons could be used to identify individual plants, as is true for fingerprints of persons, but this is not necessarily the case.

|
Technical Addendum
In order to isolate DNA from large-leafed, evergreen rhododendrons, several modifications to the isolation technique used in Rayburn et al. (1993) were made. The technique described below is taken from Iqbal et al. (1995). Three grams of leaf material were ground into a very fine powder in liquid nitrogen. The ground leaf material was then transferred into a 50 ml centrifuge tube and 15 ml of hot (65°C) 2X CTAB (2%CTAB) (i.e., cetyltrimethylammonium bromide), 1.4M NaCI, 20 mM EDTA, 0.1 M Tris-HCI (pH 8.0), 1% PP(i.e., polyvinylpyrolidon), 1% 2-mercaptoethanol) was added. The mixture was incubated at 65°C for 30 min. with occasional swirling. Fifteen ml of chloroform: isoamyl alcohol (24:1) was added, the mixture shaken, and centrifuged at 4000 rpm for 10min. The top phase was removed and re-extracted with an equal volume of the chloroform: isoamyl alcohol. The aqueous phase was removed and the DNA was precipitated with 0.6 volume of isopropanol. The DNA was pelleted at 4000 rpm for 2 min. and supernatant was discarded. After adding 10 ml of 70% ethanol to the DNA pellet, it was washed in 10% ethanol and then centrifuged at 4000 rpm for 2 min. The wash step was repeated once. The pellet was air dried (20 min.) and re-suspended in 0.5 ml of 0.1XTE. After treatment with RNase, the DNA concentration was measured using a UV-VIS spectrophotometer.
The fingerprints in this study were produced according to the procedure reported in Iqbal et al. (1995). The fingerprint of each plant is compared to all other fingerprints available. If a fingerprint has a DNA fragment at a specific site, it is given the value 1; if the fingerprint does not have a band at the site it is given the value 0. A PC SAS (SAS Institute Inc.) program (described by Mumm & Dudley, 1 994) was used to estimate the genetic distance between the rhododendron species and a phenogram created representing the genetic relationship among the various species.
Acknowledgments
About 30 plants were donated by Weldon E. Delp, who made the completion of this project very much faster and easier. (Some of these plants were also used for a more technical article already published.) The vireya was a gift from J.W. Gerdemann. The
R. smirnowii
x
R. yakushimanum
was originally obtained from Stan Hall, a cutting from a plant he propagated a number of years ago. Other leaves used for this experiment are from plants from the garden of D.W. Paden, most of which were purchased from commercial nurseries. The tissue culture material also was furnished by Paden. The University of Illinois Photographic Service prepared the figures accompanying this article.
The research reported here was supported, in part, by a grant from the ARS Research Foundation.
References
Anonymous. Genus rhododendron acquires a new species. J. Amer. Rhod. Soc.45 (2): 95; 1991.
Iqbal, M.J.; Paden, D.W.; Rayburn, A.L. Clonal stability of RAPD markers in three rhododendron species. J. Environ. Hort. 13: 43-46; 1995.
Krebs, S.L. Enzyme fingerprinting of rhododendron cultivars. J. Amer. Rhod. Soc. 49 (4): 210-215; 1995.
Kron, K.A.; Judd, W.S. Phylogenetic relationships within the rhodoreae (Ericaceae) with specific comments on the placement of ledum. Systematic Botany 15 (1): 57-68; 1990.
Mumm, R.H.; Dudley, J.W. A classification of 148 U.S. maize inbreds: 1. Cluster analysis based on RFLPs. Sci. 34: 842-85; 1994.
Paden, D.W. Rhododendron x Ledum groenlandicum , a promising innovation. J. Amer. Rhod. Soc. 28 (4): 222-223; 1974.
Rayburn, A.L; Iqbal, M.J.; Paden, D.W. Positive identification of rhododendron through DNA fingerprinting. J. Amer. Rhod. Soc. 47 (3): 137-138; 1993.
Rouse, J.L.; Knox, R.B.; Williams, E.G. Inter- and intraspecific pollination involving rhododendron species. J. Amer. Rhod. Soc. 47 (1): 23-28; 1993.
Salley, H.E.; Greer, H.E. Rhododendron hybrids (Second edition). Portland, OR: Timber Press; 1992.
Donald W. Paden is Professor Emeritus of Economics and A. Lane Rayburn is Associate Professor of Cytogenetics at the University of Illinois at Urbana-Champaign. M. Javed Iqbal is Scientific Officer, National Institute for Biotechnology and Genetic Engineering, Faisalabad, Pakistan.
