JARS v52n1 - DNA Changes Accompanying the Evolution of Plant Species
DNA Changes Accompanying the
Evolution of Plant Species
Dr. Benjamin D. Hall
Chairman, ARS Research
Committee
Department of Botany, University of Washington
Seattle, Washington
Rhododendron species: Why are there so many of them? What is the operational definition of a species? On what continent did rhododendrons first come to exist? How long ago did the common ancestor of R. macrophyllum and R. maximum exist? Many members of this society who observe, admire and grow wild rhododendron species find these questions intriguing. Because of recent developments in DNA technology, it now appears possible that definitive answers to these and other questions and about the family tree (phylogeny) of the different Rhododendron species may soon be forthcoming.
An interesting theoretical explanation for several aspects of rhododendron species formation (speciation) was put forward by Irving and Hebda in this Journal (Irving and Hebda, 1993). They emphasized the role of physical barriers, such as very high mountain ridges and glaciers, in preventing the exchange of pollen (and genes) between different localized populations of the same species. Equally effective barriers to such exchange can come about through differences in flowering season or in pollinator preference.
Over times as long as one million years, reproductive isolation inevitably leads to genetic differences between isolated populations, through mutational change and the prevention of crossbreeding. Speciation results from such isolation and differentiation. Irving and Hebda made further suggestions to explain why so very many species are found in China, the Himalayas and Southeast Asia, where uplifting, glaciation and erosion have led to a large number of isolated rhododendron populations.
The remainder of this article will focus on the genetic changes accompanying speciation and evolution: changes driven in part by natural selection and in part by strictly random events. Related articles by Kathleen Kron and Amy Denton in the next two issues of the Journal will describe how studies of Rhododendron DNA make it possible to determine the relatedness of different rhododendron species with considerable certainty and even to reconstruct plausible scenarios describing how these species have diverged from one another and have been dispersed across the Northern Hemisphere.
Our understanding of species formation and the genetic basis for it derives from the work of two 19th century biologists, Charles Darwin and Gregor Mendel, considered by many the two greatest biologists of all time. Darwin's concept of evolution through natural selection and Mendel's elucidation of the gene as the unit of inheritance were combined in the 1930s and 1940s to provide a clear quantitative explanation of speciation. Several mathematically-oriented geneticists and evolutionists in Britain and the United States addressed the following questions: "Suppose two initial populations of an organism that are identical in their genes become isolated by some barrier and suppose that both populations survive for many eons. Will they become different and, if so, why? What are the driving forces?" One driving force certainly would be natural selection. If environmental changes ensue after population A becomes separated from population B, for example, if one locale is drier or colder than the other, then those exceptional genetic types that adapt better to that particular environment will gradually take over each population. Solar radiation damage to DNA and occasional errors in DNA duplication provide a continuous supply of new genetic types upon which natural selection may act.
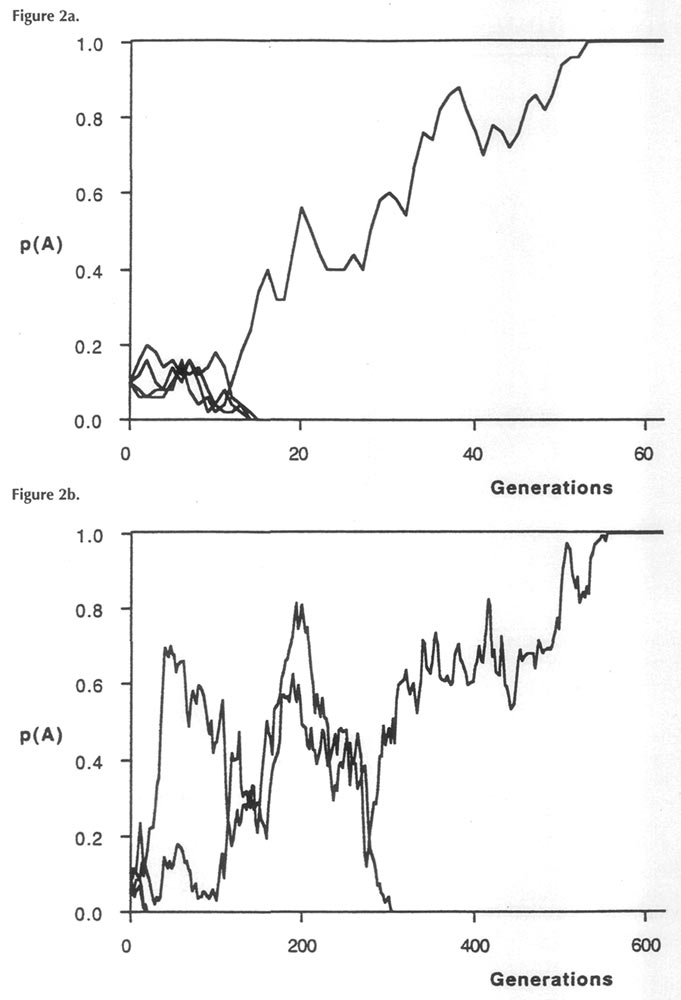
Twentieth century geneticists identified a second major contributing factor to the evolutionary divergence between isolated populations; this is known as random genetic drift. Drift occurs because the passing on of genes from one generation to the next depends upon many different random events. In the case of a new genetic type of Rhododendron arising by a chromosomal mutation, its chance of reproduction is affected by nutritional control of flowering (climate, shade, soil); proximity to another plant, encounters with bumblebees, etc. Other chance events will affect dissemination and germination of the seed. Compared to the normal genetic types of rhododendrons surrounding it, the mutant type may either be lucky or unlucky at siring progeny. With a relatively few lucky breaks, a new genetic type can quickly displace the original type from a small isolated population, especially if the mutation confers a selective advantage to the novel genetic type. Figure 2 illustrates a computer simulation of genetic drift for two hypothesized rhododendron populations. The population in Fig. 2a contains four exceptional (mutant) genes within a population of 25 plants. Because each plant is a diploid (formed by pollen fertilizing ova each of which has one set of genes), there are two gene copies per plant, making a total of 50. When these plants are allowed to mate and reproduce randomly (by computer simulation), three mutant copies are quickly eliminated by drift (Fig. 2a), while the fourth one manages to take over the population by about the sixtieth generation. Fig. 2b shows a similar computer simulation for a larger population (100 plants), also containing four new mutant types. In this case, a mixture of original and mutant types continues to exist over many more generations (at least 200). The difference between the outcomes in these two cases illustrates the strong effect of population size upon the rate of genetic drift - the basis for the large effect of reproductive isolation upon species differentiation.
|
|---|
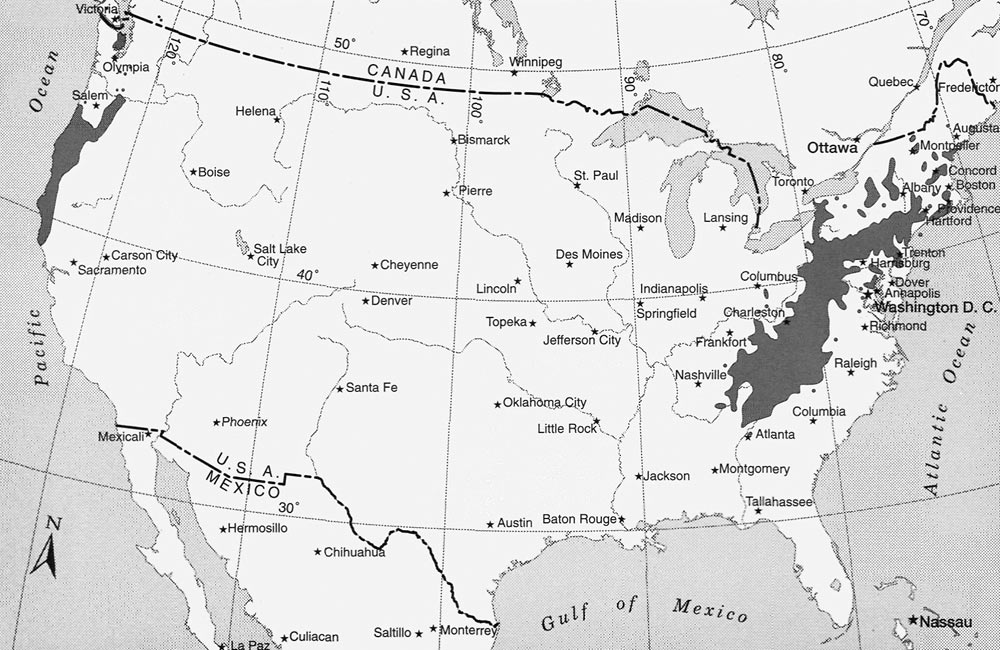
Figure 1. Natural ranges of Rhododendron macrophyllum on the west coast and Rhododendron maximum on the east coast of the United States. Map courtesy of the Western North American Rhododendron Species Project . |
The process of change in genetic composition, shown in Figure 2 for one gene undergoing a change in frequency within a single population, would occur independently within any two isolated populations even if they shared a common ancestor. It is the overall effect of many such mutational changes, in each one of more than 10,000 different genes, that constitutes the genetic difference in DNA between two related species, for example those between Rhododendron maximum and Rhododendron macrophyllum . All other things being equal, the longer ago the two species became reproductively isolated, the more their genes will differ. The essential nature of these differences is a permanent, heritable change in the structure of DNA. The variable components of DNA, called bases, are four in number and are denoted by the shorthand symbols A, T, G and C. These four bases are attached to an unvarying central core of the DNA double helix which is called the backbone. What gives any region of DNA its individuality is the sequence of bases attached to the backbone. For example, the sequence GACCTG is one which can specify two corresponding amino acids (Aspartic acid • Leucirie) in a protein. Mutations are changes in DNA base sequence which, generally speaking, cause protein changes and changes in the appearance or the function of an organism.
|
|---|
Figure 2a & b. Computer simulations of successive generations for two Rhododendron populations, in each of which 4 mutant genes exist initially and within which random matings occur, a) Population size = 25; b) Population size = 100. On the g-axis is plotted frequency of each mutant type. |
The most common type of mutation observed is a base substitution, such as the change GACCTG -» GACGTG within the DNA of a gene. We refer to this mutation as a C to G substitution. When the same gene is compared between widely different species, differences in the mutational history of the two species are reflected by multiple differences in their DNA sequences, as in Figure 3, which compares the base sequences for a short region of the same gene isolated from Rhododendron macrophyllum , Lycopersicon esculentum , Magnolia virginianam , Nymphaea colorata , and Ginkgo biloba .
Figure 3. Comparison of Plant DNA Sequences.
Lycopersicon ATAAATGATCTTGTGCAAGCGCGCCTTAATCCTGGCGAC
Rhododendron ATAAATGATCTTACATCTGCACGTCTCTGCCCAGATGAC
Magnolia ATTTCAGATCTTATAAGTGCAAGGCACAATCCAGAAGAA
Nymphaea ATTTCAGATTTAGTCAGTGCAAGGCTCAATCCTGAGGAG
Ginkgo ATTTCGGACCTTGTGACTGCTAGAAATAATCCGGAAGAG
Molecular systematics deduces patterns of descent by using such DNA differences to measure the degree of relatedness between the same gene in different organisms and, ultimately, to determine how the organisms themselves are related to one another. (A cautionary note: such relationships are regarded as provisional and tentative until the studies are extended to other genes and until the molecular conclusions are compared with those from morphological studies.)
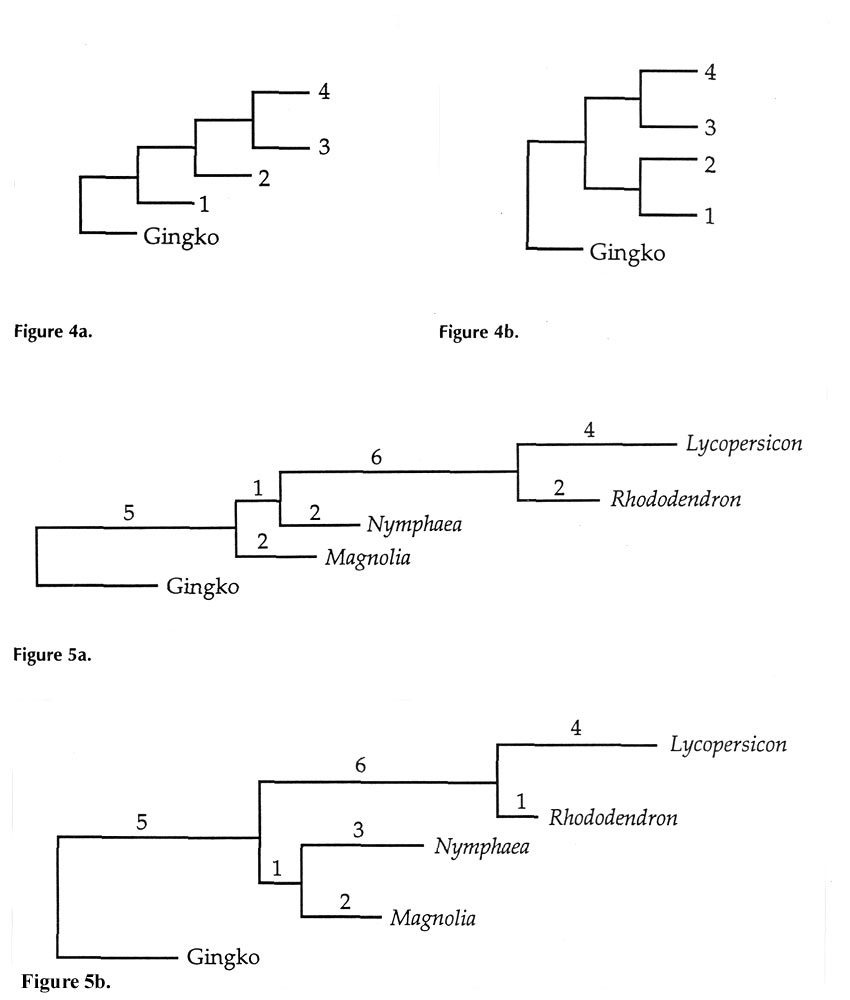
A major objective of systematics is to construct a phylogenetic tree for the group of plant or animal species being studied. The very limited data set in Figure 3 can be used to illustrate one of the most widely used procedures for tree construction, called the method of maximum parsimony. Of the five plant species listed four are angiosperms, while the Ginkgo tree belongs to the group of gymnosperms (naked seed). If we make the non-controversial assumption that the four angiosperms are descended from a common ancestor not shared with Ginkgo , this provides a sense of direction to the tree. Its base or "root" should lie between Ginkgo and all of the angiosperms. All possible trees that describe this situation have one of the two shapes shown in Figures 4a and b.
The different possible trees correspond merely to different relative positions of Lycopersicon , Rhododendron , Magnolia , and Nymphaea . Altogether, 15 different possible trees can be drawn for these four species by permuting their locations between the four positions. (Note that, in Figure 4a, positions 3 and 4 are equivalent, and in Figure 4b, not only is 3 equivalent to 4, but 1 is equivalent to 2, as well.)
In parsimony analysis, the assumption is made that the "true" tree (true defined as consistent with the most sequence data) is that which has the shortest overall length, when the lengths of all horizontal segments are added together. Each segment length is simply the number of DNA base changes that must have taken place in that portion of the evolutionary tree. The data used to assign these changes comes from the bases in bold in Figure 3, positions where two or more plants each have the same change. Numbers on the horizontal parts of Figures 5a and 5b show how many changes have occurred at these critical sites. An only slightly less parsimonious tree is one like 5a, but with Magnolia and Nymphaea exchanging places.
|
|---|
Despite its limitations, this brief exercise with the sequences of Figure 3 makes a certain amount of botanical sense. Lycopersicon and Rhododendron belong to the group of eudicots, more advanced flowering plants, whereas Nymphaea and Magnolia are less advanced in flower complexity.
If the plants studied had been much more closely related, for example, if all were members of the Ericaceae, sequences of this particular gene segment would have been uninformative. All the sequences would be nearly identical, or within the "noise level" of occasional genetic change. Given that interest in determining the tree structure for related genera within plant families is very high, how can this be done? Either of two modifications of strategy can give the desired information. One is simply to sequence a greater length of DNA, either of one or several genes. Dr. Kron's article will illustrate how sequences from several genes have helped in understanding Rhododendron phylogeny.
An alternative way to achieve greater resolution in phylogenetic analysis, rather than sequencing more DNA, is to focus upon regions of DNA that change more rapidly. Generally, these are regions of DNA that do not specify anything essential to the life processes of the organism and thus will tolerate more rapid change than the sequences in Figure 3.
In her studies of the DNA of Rhododendron species, particularly those of sections Hymenanthes and Rhododendron , Amy Denton has used this principle to resolve their pedigrees. In sequencing a non-functional DNA region among and within a number of these Rhododendron species, Dr. Denton has made a critical evaluation of such high-resolution phylogenetic analysis. Her research disclosed an unexpectedly high genetic diversity within Western native populations of Rhododendron macrophyllum , suggesting a complex and varied evolutionary history for this familiar species.
Literature Cited
Irving, E. and R. Hebda. 1993. Concerning the Origin and Distribution of Rhododendrons.
JARS 47:39.
Suggestions for Further Reading
Avise, John C. Molecular Markers, Natural History and Evolution. New York: Chapman &
Hall, 1994.
Li, W-H, and D. Graur. Fundamentals of Molecular Evolution. Sunderland, MA: Sinauer
Associates, Inc., 1991.

